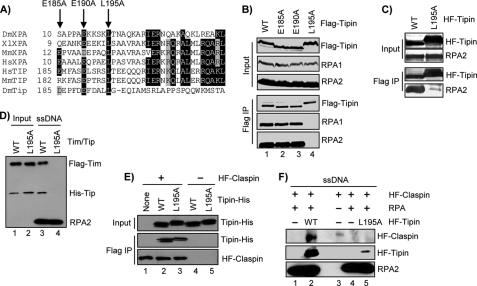
FIGURE 3.
Identification and characterization of a Tipin RPA-binding mutant. A, alignment of XPA and Tipin amino acid sequences. Black shading shows amino acid identity, and gray shading highlight indicates similarity. B, FLAG-tagged forms of Tipin were transiently expressed in HEK293T cells, immunoprecipitated with anti-FLAG-agarose, and analyzed by SDS-PAGE and immunoblotting with antibodies against FLAG, RPA1, and RPA2. Note that mutation of these amino acids alters Tipin mobility on SDS-PAGE. C, His-FLAG-tagged Tipin (HF-Tipin; WT and L195A) from baculovirus-infected insect cells was immobilized on anti-FLAG-agarose and then incubated with RPA. Resin was washed, and bound proteins were analyzed by SDS-PAGE and immunoblotting. D, FLAG-Timeless/His-Tipin (Tim/Tip) complexes prepared by baculoviral co-infection and anti-FLAG-agarose purification were incubated with immobilized ssDNA containing saturating amounts of RPA. Bound proteins were analyzed by SDS-PAGE and immunoblotting. Input represents 50% of the Timeless-Tipin complex used in the binding reaction. E, His-FLAG-tagged Claspin (HF-Claspin) from baculovirus-infected insect cells and immobilized on anti-FLAG resin (lanes 1–3) was incubated overnight at 4 °C with 1.5 μg of either Tipin-His-WT or Tipin-His-L195A purified from E. coli in 100 μl of binding buffer. Anti-FLAG resin lacking Claspin was used as a negative control (lanes 4–5). Input represents 5% of the Tipin used in the binding reaction. F, immobilized 80-mer ssDNA lacking or containing saturating amounts of RPA was incubated in reactions with HF-Claspin alone or together with HF-Tipin-WT or HF-Tipin-L195A. The beads were washed, and bound proteins were analyzed by SDS-PAGE and immunoblotting. IP, immunoprecipitation.