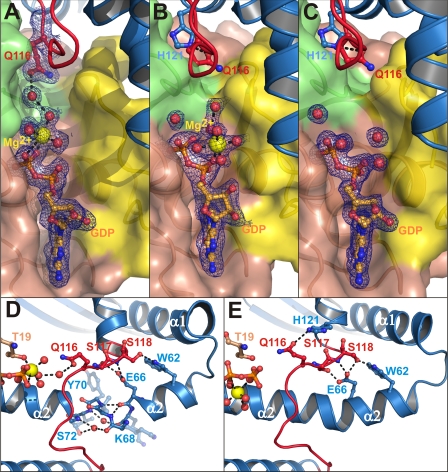
FIGURE 3.
Nucleotide binding site and catalytic loop region in IpgB2·RhoA structures. IpgB2 is shown as a blue schematic, with the catalytic loop marked in red. GDP is shown as orange balls and sticks, and Mg2+ and water molecules are shown as yellow and red spheres, respectively. A, GDP-binding pocket and interaction of IpgB2 residue Gln116 in complex A. The [2Fo − Fc] electron density map contoured at 1 σ for GDP, Mg2+, the Mg2+ coordination sphere, and Gln116 is colored as blue mesh. B and C, similar representation as in A for complexes B and C, respectively. For A–C, RhoA is shown as a surface representation, with switch I, switch II, and other regions of RhoA as yellow, green, and brown surfaces, respectively (compare also supplemental Fig. 2 for a more detailed view in stereo representation). D, helix α2 in complex A, showing a conformation that is disrupted by two water molecules. Such a disrupted helix conformation is not observed in crystals of complex B (E) or of complex C (not shown). RhoA residues are shown as brown balls and sticks in D and E.