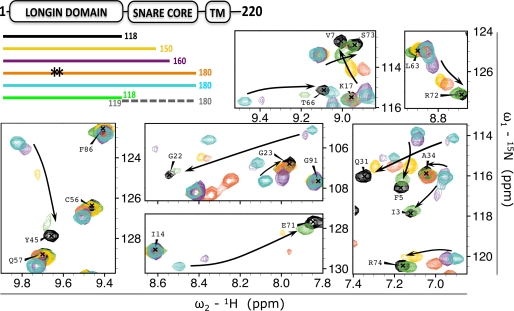
FIGURE 4.
Effect of sequence specificity, construct length, and covalent linkage on the LD-SNARE core interaction. The effect of truncation and mutation of 15N-labeled VAMP7 constructs on LD resonances is shown as follows: VAMP7(1–118) (black), VAMP7(1–150) (yellow), VAMP7(1–160) (purple), VAMP7(1–180)-L43S/Y45S (orange), VAMP7(1–180) (cyan), and VAMP7(1–118) (green) incubated with non-labeled VAMP7(119–180) (dashed gray line). The color-coded bar diagram in the top left corner indicates the boundaries of these six constructs for which the spectra are shown with respect to the domain architecture of full-length VAMP7, residues 1–220: LD, SNARE core domain, and transmembrane domain (TM). Black stars on the orange bar represent the L43S/Y45S mutation from the canonical amino acid sequence of VAMP7. An overlay of the 1H{15N}-HSQC spectra of these proteins is shown with matching colors in seven representative regions of the spectra (same regions shown in Fig. 3, A and C). The three panels on the left are shown with lower contour levels to display broadened resonances. Black arrows indicate the chemical shift perturbation trends from the closed to the open conformation.