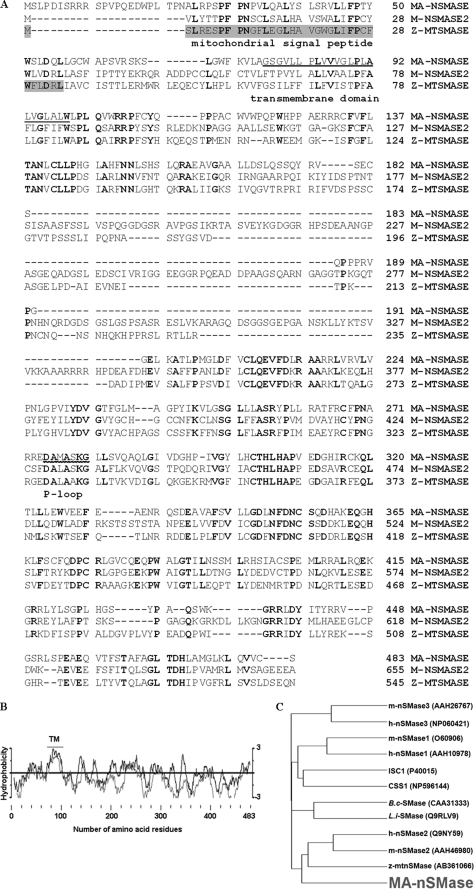
FIGURE 1.
Sequence analysis of mouse MA-nSMase. A, alignment of the deduced amino acid sequences of mouse MA-nSMase, mouse nSMase2, and zebrafish mitochondrial SMase (Z-MTSMASE). The sequences were aligned by the GCG Pileup program. Identical residues in all the three sequences are indicated by bold characters. The mitochondrial signal peptide in zebrafish mitochondrial SMase is highlighted. The predicted transmembrane domain and p-loop like domain are underlined. B, hydrophobicity profile of mouse MA-nSMase. The deduced amino acid sequence of MA-nSMase was analyzed by the method of Kyte-Doolittle (dark line) and by the Goldman method (light line) for hydrophobicity plotting. The predicted transmembrane (TM) domain is indicated. C, evolutionary relationships between MA-nSMase and other nSMases. A phylogenetic tree of various nSMases was plotted by the GCG program using the Kimura protein distance correction. The length of each horizontal line in the tree is proportional to the difference of the amino acid sequences. The nSMase sequences are from human (h), mouse (m), Bacillus cereus (B.c) and Listeria ivanovii (L.i). The ISC1 and CSS1 sequences are from Saccharomyces cerevisiae and Schizosaccharomyces pombe.