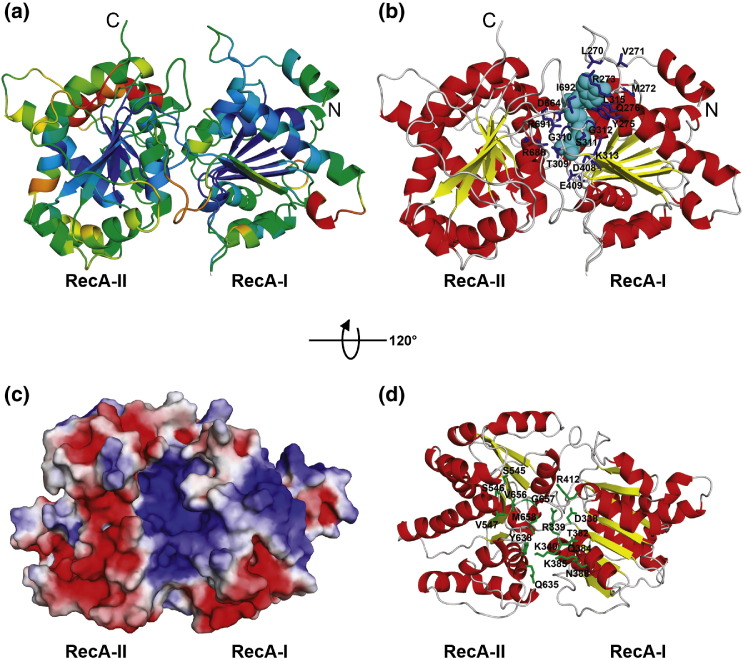
Figure 7.
Model of the central domain of EcoR124I HsdR. (a) Model coloured according to MetaMQAP evaluation (well-scored regions are coloured blue, poorly scored regions are coloured red). (b) Active site of the central domain: ATP moiety (cyan) and magnesium ion (pink) coordinates copied from 2db3 structure. The residues predicted to be involved in ATP-binding (shown in blue) are located in a cleft between two NTPase domains. The model is coloured according to secondary structure (red, helices; yellow, β-strands; grey, loops). (c) Electrostatic potential mapped onto the molecular surface of the central domain (positively and negatively charged regions are coloured in blue and red, respectively). (d) Putative DNA-binding residues (coloured green).