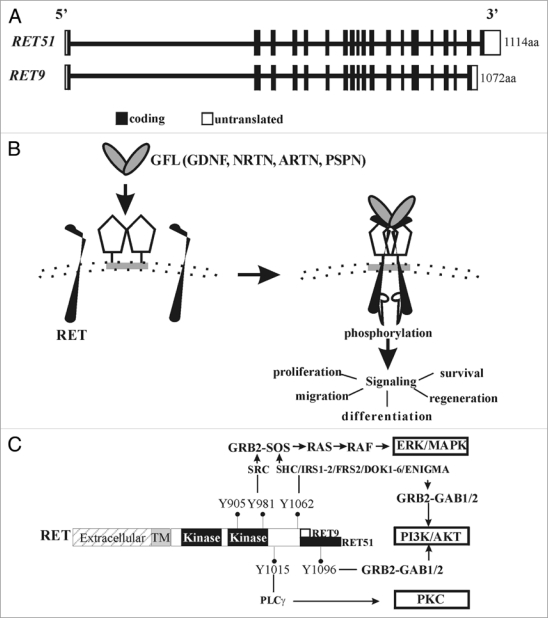
Figure 1.
Ret gene structure and signaling. (A) Gene structure of RET. RET is located at chr 10q11.2 and consists of 20 exons (rectangular boxes). Depicted are two major RET isoforms, RET51 and RET9, generated by alternative splicing. (B) Diagram showing RET activation by glial cell line-derived neurotrophic factor (GDNF) family ligands (GFLs). A dimer of one of the four GFLs (GDNF; Neurturin, NRTN; Artemin, ARTN; Persephin, PSPN) associates with homodimers of one of the four GPi-linked corereceptor's (GFRα1–4) to form multimeric complex with a RET homodimer. This leads to activation of RET kinase domain in the cytoplasmic side by phosphorylation, and downstream signaling cascades that regulate the indicated biological responses. (c) Gross anatomy of RET. Different RET domains are shown, extracellular, transmembrane (TM) and cytoplasmic domain that harbors the kinase activity regions. Key RET tyrosine (Y) residues, the preferred intracellular adaptors that they bind to, and the downstream signaling cascades that are activated are shown. Y905 is a residue important for RET kinase activity. Note the extra Tyr Y1096 in the RET51 isoform directly binds to GRB2 to preferentially activate PI3K/AKT pathway. Thus, both RET-Y1062 and RET-Y1096 can activate PI3K through GRB2-GAB1/2.