Figure 4. Protein architecture of the Saccharomyces cerevisiae kinetochore.
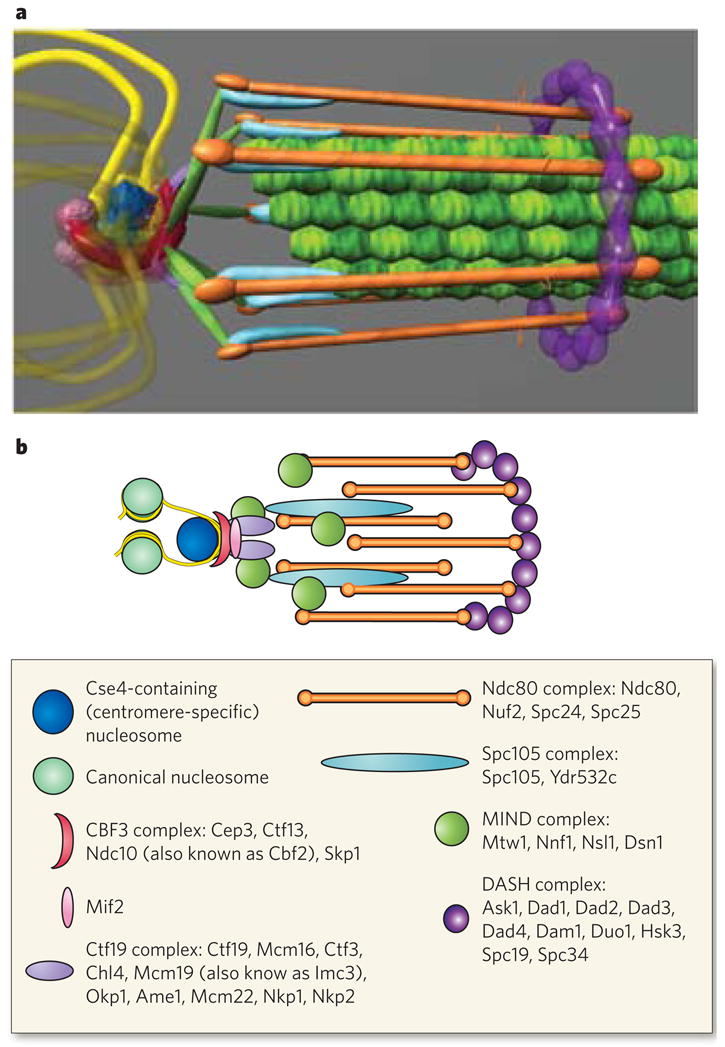
The structure of the kinetochore is depicted in two ways. a, This structure reflects the average positions of kinetochore proteins within the S. cerevisiae kinetochore measured by invivo super-resolution microscopy75. The positions of microtubule-binding proteins with respect to the microtubule plus end are key to understanding the mechanism of force generation at the kinetochore. The interface between the microtubule-binding side of the kinetochore and the chromatin-binding side in S. cerevisiae, as well as in multicellular eukaryotes, remains poorly characterized93. From the left, the DNA (yellow strands) is wrapped in a positive supercoil around a histone core that contains the centromere-specific H3 variant (dark blue). The DNA-binding region of the kinetochore is formed by the CBF3 complex (dark pink). An additional DNA-binding protein, Mif2, is shown proximal to centromeric DNA (light pink sphere). From the right, the microtubules (green filaments) are surrounded by the DASH complex (purple ring), whose constituents are abundant enough to form a ring. The main microtubule-binding sites within the kinetochore are in the Ndc80 complex (orange rods). There are several linker complexes: the Spc105 complex is depicted in light blue, and the MIND complex in green; the Ctf19 complex is shown as small purple linkers between the CBF3 and MIND complexes. b, This illustration is a schematic representation of the structure in a. Data are based on the hydrodynamic properties, protein number and spatial position of the members of each protein complex. The structure assumes a symmetrical arrangement of kinetochore protein complexes around the microtubule lattice (green in a). The known components of the various protein complexes are listed in the box.
