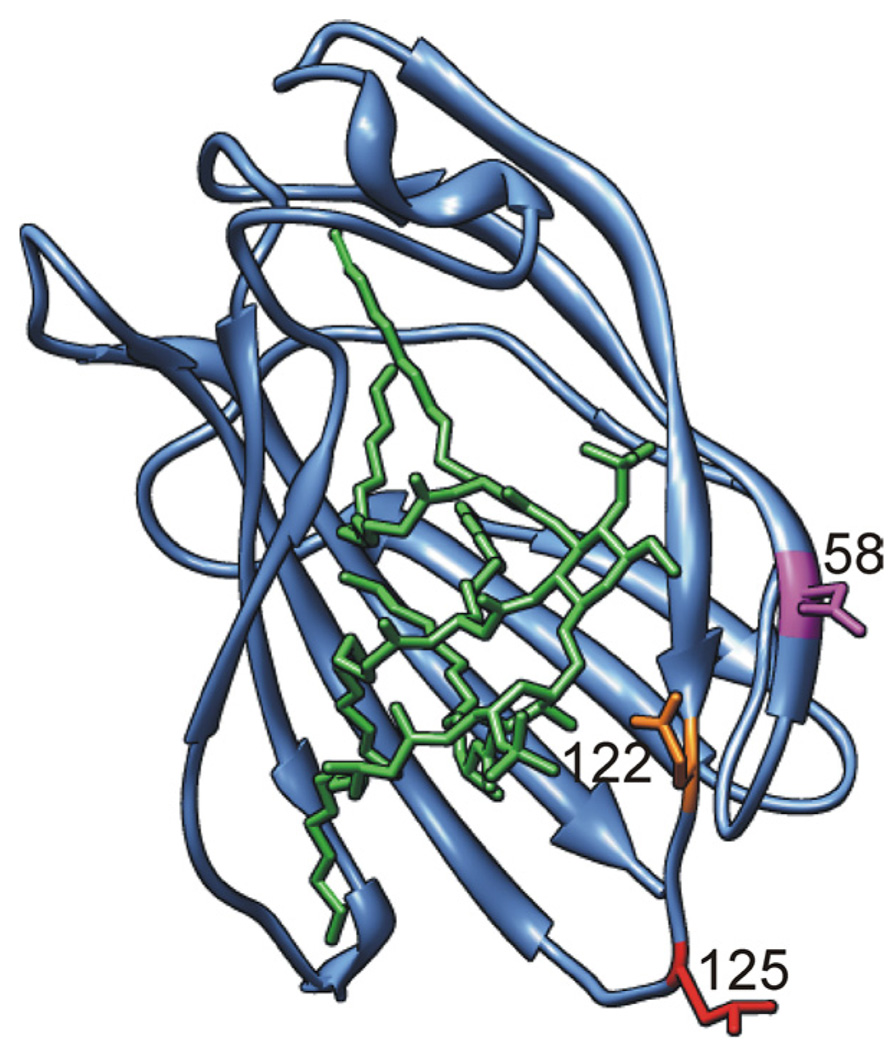
FIGURE 1.
Differences in charged amino acid residues at the rim of the LPS binding pocket of human vs murine MD-2. Ribbon model of the three-dimensional structure of mMD-2 with side chains of residues N58, E122, and L125 shown in magenta, orange, and red, respectively (lysines in hMD-2). Lipid A is shown as a stick model in green. Model was prepared by UCSF Chimera (40) based on the coordinates of mMD-2 (PDB code 2Z64) with lipid A docked from the TLR4 · MD-2 · Ra-LPS complex (PDB code 2FXI) (20, 22).