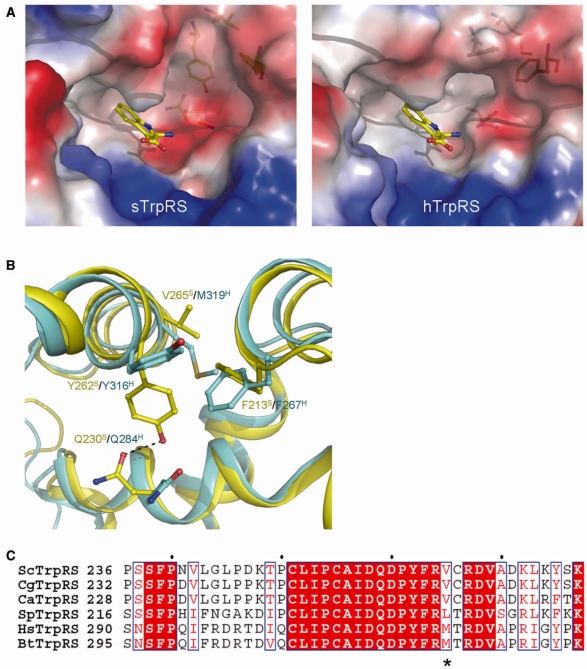
Figure 4.
Conformational differences of the Trp-binding subsite between the apo sTrpRS and hTrpRS structures. (A) Electrostatic surfaces of the Trp-binding pockets in the apo sTrpRS (left panel) and hTrpRS (right panel) structures. A tryptophan molecule is docked to show the Trp-binding subsite. (B) Structural comparison of the Trp-binding subsite of the apo sTrpRS (yellow) with that of the apo hTrpRS (cyan). The residues responsible for the structural differences are labeled and shown with stick-and-ball models. The sequence variance at position Val265 S/Met319H is a key for the structural differences of the Trp-binding subsite. (C) Sequence alignment of sTrpRS with TrpRSs from other yeasts including C. glabrata, C. albicans and S. pombe, and with those from higher eukaryotes including H. sapiens and B. taurus. The position equivalent to that of Val265S/Met319H is marked by an asterisk. The sequence alignment is generated by ESPript (58).