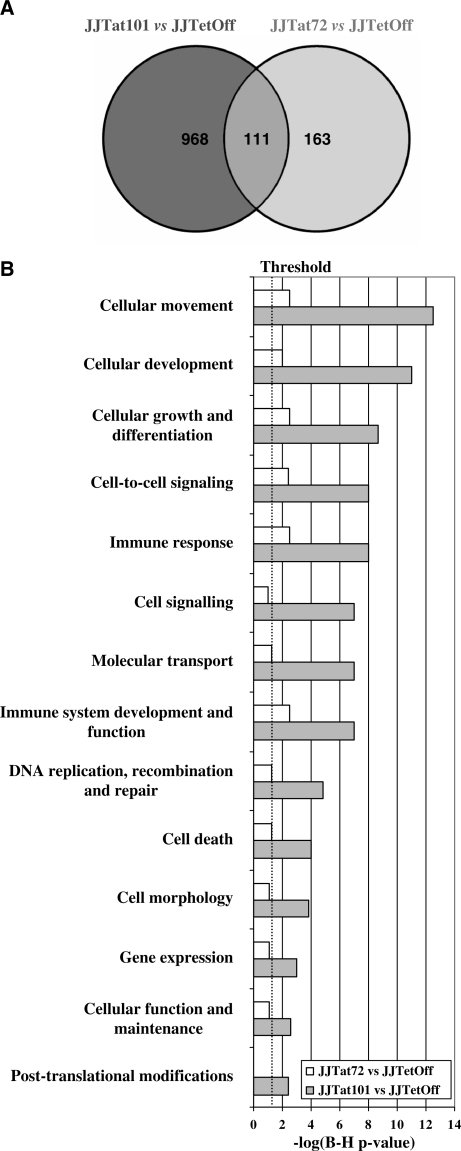
Figure 4.
Gene expression and bioinformatics analysis. (A) Gene expression patterns of Jurkat-Tat101 and Jurkat-Tat72 were compared with control cells. Data were considered statistically significant when q-value was <5% and fold change >2 or <−2. These filtering criteria yielded 1079 analysable gene spots for Jurkat-Tat101 versus control, and 274 analysable gene spots for Jurkat-Tat72 versus control. These results are summarized in a Venn diagram. The overlapped area represents 111 genes that were deregulated in both cell lines. (B) Analysis of functional and canonical pathways for specific gene datasets generated with the B-H Multiple Testing Correction p-value determined that all cellular functions were mostly altered in Jurkat-Tat101. Data representing the −log(B–H p-value) are shown in a histogram for Jurkat-Tat101 versus control and Jurkat-Tat72 versus control comparisons.