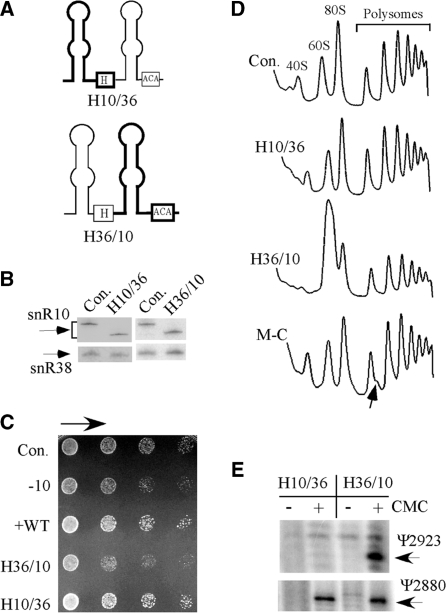
Figure 4.
The processing function of snR10 is mediated by its 5′ hairpin. (A) The roles of the 5′ and 3′ hairpins in snR10 function were examined with hybrid snoRNAs containing hairpins from snR10 and snR36 in test cells lacking snR10. The structures of the hybrid snoRNAs are depicted schematically, with the snR10 portion shown with thick lines and the snR36 portion with thin lines. (B) The hybrid snoRNAs are expressed at normal levels as evident in northern hybridization results. (C) Normal growth occurred with the hybrid containing the 5′ hairpin of snR10 (H10/36), but not the hybrid containing the 3′ hairpin of snR10 (H36/10). The snoRNA snR38 was used as a control for loading. (D) A normal ribosome–polysome pattern was restored by the hybrid snoRNA with the 5′ hairpin from snR10 (H10/36). Half-mer polysomes are evident in test cells that express a snR10 mutant lacking a C nucleotide (M-C) in the guide domain; this deletion blocks Ψ2923 formation (12). Accumulation of half-mer polysomes is indicated by an arrow. (E) The Ψ2923 modification is formed with a hybrid snoRNA containing the 3′ hairpin of snR10 (H36/10). Total RNA was treated with CMC and Ψ detected by primer extension analysis. The extension stops representing Ψ2923 and Ψ2880 (guided by snR10 and snR34, respectively) are identified.