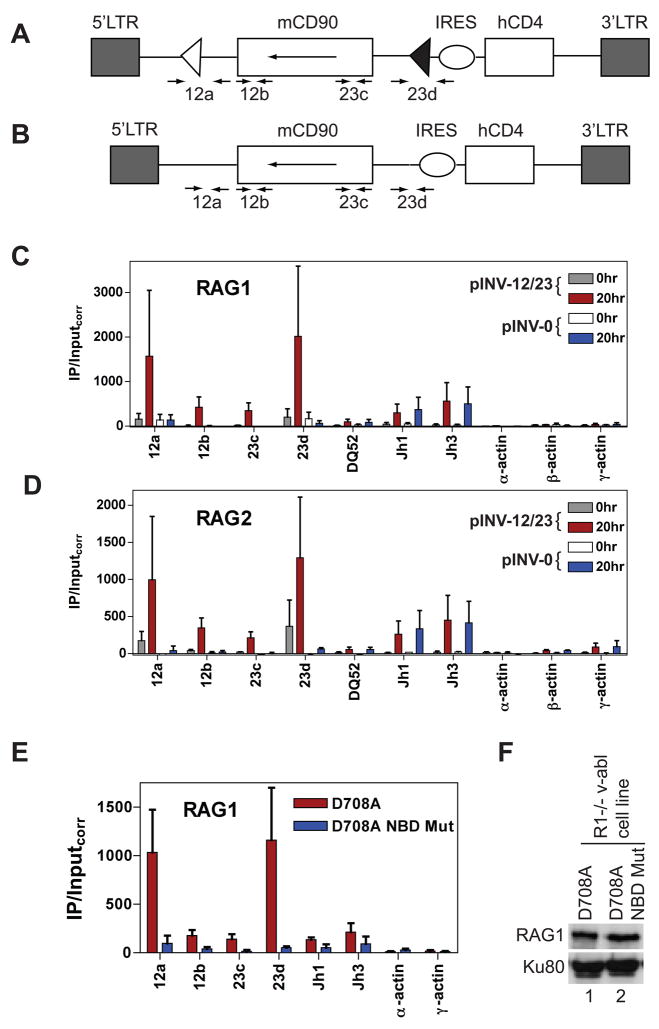
Figure 5. The RSS and nonamer binding domain are important for RAG binding.
(A and B) The pINV-12/23 (A) and pINV-0 (B) recombination substrates. The 5′ and 3′ long terminal repeats (LTR), mouse CD90 gene (mCD90) encoding Thy1.1, and human CD4 gene (hCD4) are indicated as rectangles, the internal ribosome entry site (IRES) as an oval, and the 12RSS and 23RSS as white and black triangles, respectively. The mCD90 gene lies in opposite transcriptional orientation (long arrow) to that of transcription originating in the 5′LTR. The positions of PCR primers are indicated with short arrows. Primer pairs 12a and 23d span the 12RSS and 23RSS respectively in pINV-12/23, and generate 109 bp (12a) and 61 bp (23d) smaller products with pINV-0 than with pINV-12/23 due to deletion of the RSSs. Primer pairs 12b and 23c lie 69 bp 3′ of the 12RSS and 91 bp 5′ the 23RSS, respectively. Not drawn to scale.
(C-D) Binding of RAG1 (C) or RAG2 (D) were assessed by ChIP in the D345 v-abl transformed cell line infected with pINV-12/23 (gray and red bars) or pINV-0 (white and blue bars) either prior to RAG induction (0hr) or after 20 hours of RAG induction (20hr) at the gene segments or regions indicated. Data are the average of two independent experiments and are presented as in Fig. 1. See also Fig. S6.
(E) Binding of RAG1 was assessed in a R1−/− cell line containing pINV-12/23 and infected with a retrovirus expressing D708A RAG1 (red bars) or nonamer binding domain (NBD) mutant D708A RAG1 (blue bars) and a linked blastocydin resistance gene. Data were collected from blastocydin-resistant clones 20 hours after treatment with STI-571 and are the average of two independent experiments. Similar data were obtained using a second set of independently derived clones (data not shown).
(F) Western blot of D708A RAG1 and NBD-mutant D708A RAG1 proteins in infected R1−/− v-abl cells. A separate blot of the same protein extracts was probed for Ku80 to confirm equal loading.