Figure 5.
Targets of Less Conserved miRNAs Are Difficult to Identify and Inconsistent between A. thaliana and A. lyrata.
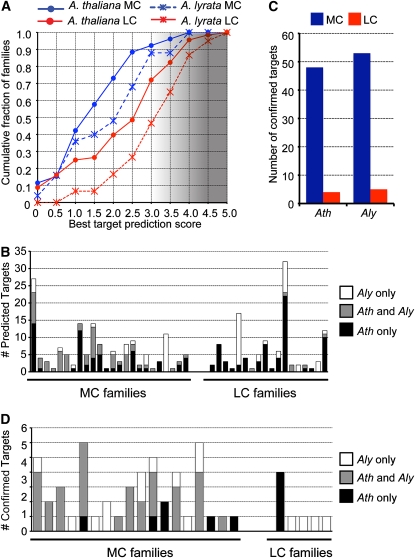
(A) Cumulative distributions of the number of miRNA families with the indicated target prediction scores. The lowest scoring prediction (i.e., the most confident prediction) for each family was used. MC, more conserved; LC, less conserved. The gradient of shading indicates increasingly less confident predictions, beginning at a score of 3.
(B) miRNA target predictions by family. The number of predicted targets found only in A. thaliana (Ath), only in A. lyrata (Aly), or syntenic homologs predicted in both species are shown. Families without any predicted targets in either species are omitted, as are families that were expressed only in a single species.
(C) Sliced targets confidently found by degradome sequencing. Sliced targets were those that were found in both biological replicate degradome libraries for the given species.
(D) As in (B) for degradome-confirmed targets.