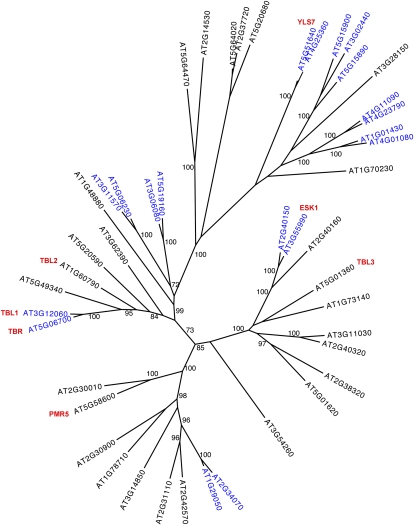
Figure 5.
Unrooted, bootstrapped tree of the TBL gene family in Arabidopsis. MAFFT (version 5) was used to create an alignment of the protein sequences available at TAIR. Highly variable, gapped N-terminal regions were removed, and a maximum likelihood phylogeny was generated with RAxML model PROTCATWAG (Stamatakis, 2006) and bootstrapped (n = 1,000 trials) to generate the final tree. Bootstrap values greater than 70 are shown. Segmental/tandem gene pairs are highlighted in blue, and genes mentioned in the text are shown in red.