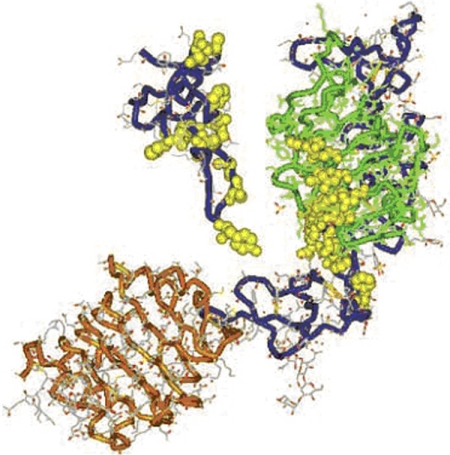
Fig. 1.
Structures of the IGF-I receptor and its ligand. Three-dimensional structure of large domain 1 (L1)–Cys-rich (CR)-L2 domain of IGF-IR determined by X-ray crystallography. An extended bilobed structure (40 × 48 × 105 Å) comprises the two globular L-domains with a new type of right-handed β-helix fold that flanks the CR domain. They seem to be part of the leucine-rich-repeat superfamily. Although L1 (residues 1–15; green) contacts the CR domain (blue) along its length, there is minimal contact with L2 (residues 300–460; orange). Flexibility between CR domain and L2 could affect ligand binding. A −30-Å diameter cavity represents a potential binding pocket. The amino acids that have been determined by alanine-scanning mutagenesis to be important for ligand binding are shown in yellow as van der Waals spheres. Three-dimensional IGF-I structure is based on the X-ray coordinates from Brzozowski et al. (2002). The backbone is shown in blue. [Reproduced from De Meyts P and Whittaker J (2002) Structural biology of insulin and IGF1 receptors: implications for drug design. Nat Rev Drug Discov 1:769–783. Copyright © 2002 Nature Publishing Group. Used with permission.]