Abstract
Tomatoes in Guatemala have been affected by a new disease, locally known as “mancha de chocolate” (chocolate spot). The disease is characterized by distinct necrotic spots on leaves, stems and petioles that eventually expand and cause a dieback of apical tissues. Samples from symptomatic plants tested negative for infection by tomato spotted wilt virus, tobacco streak virus, tobacco etch virus and other known tomato-infecting viruses. A virus-like agent was sap-transmitted from diseased tissue to Nicotiana benthamiana and, when graft-transmitted to tomato, this agent induced chocolate spot symptoms. This virus-like agent also was sap-transmitted to Datura stramonium and Nicotiana glutinosa, but not to a range of non-solanaceous indicator plants. Icosahedral virions ~28–30 nm in diameter were purified from symptomatic N. benthamiana plants. When rub-inoculated onto leaves of N. benthamiana plants, these virions induced symptoms indistinguishable from those in N. benthamiana plants infected with the sap-transmissible virus associated with chocolate spot disease. Tomatoes inoculated with sap or grafted with shoots from N. benthamiana plants infected with purified virions developed typical chocolate spot symptoms, consistent with this virus being the causal agent of the disease. Analysis of nucleic acids associated with purified virions of the chocolate-spot-associated virus, revealed a genome composed of two single-stranded RNAs of ~7.5 and ~5.1 kb. Sequence analysis of these RNAs revealed a genome organization similar to recently described torradoviruses, a new group of picorna-like viruses causing necrosis-associated diseases of tomatoes in Europe [tomato torrado virus (ToTV)] and Mexico [tomato apex necrosis virus (ToANV) and tomato marchitez virus (ToMarV)]. Thus, the ~7.5 kb and ~5.1 kb RNAs of the chocolate-spot-associated virus corresponded to the torradovirus RNA1 and RNA2, respectively; however, sequence comparisons revealed 64–83% identities with RNA1 and RNA2 sequences of ToTV, ToANV and ToMarV. Together, these results indicate that the chocolate-spot-associated virus is a member of a distinct torradovirus species and, thus, another member of the recently established genus Torradovirus in the family Secoviridae. The name tomato chocolate spot virus is proposed.
Introduction
In 1999, a new disease was observed in tomato fields in Guatemala, including the Salama Valley and other production areas. Leaves of infected plants initially showed epinasty and mottling, but then developed necrotic spots, especially in the basal portion of newly emerging leaves. Eventually, the necrotic symptoms spread to stems and petioles, sometimes resulting in dieback of entire shoots (Fig. 1a, b, c and d). Fruits were relatively small but generally did not show necrosis or any other obvious symptoms. Locally, the disease was referred to as “mancha de chocolate” (chocolate spot). It was noted that the disease tended to occur at higher elevations and was consistently associated with high populations of the greenhouse whitefly (Trialeurodes vaporariorum). Although chocolate spot disease symptoms showed some similarity to those induced by tomato spotted wilt virus and tobacco streak virus, tests for these and other known tomato-infecting viruses were negative.
Fig. 1.

Chocolate spot disease symptoms in tomato in Guatemala and in tomatoes inoculated with sap or grafted with shoots from N. benthamiana plants infected with purified virions of the chocolate-spot-associated virus. a Close-up showing development of necrotic spots in basal portions of leaves. b Typical necrosis symptoms in the basal portion of leaves, and in petioles and stems. c Crumpled and necrotic leaves, and necrosis of petioles and stems. d Necrosis of stems and petioles, resulting in dieback of an entire shoot. e The development of necrotic spots in basal portions of leaves of tomato cv. El Senor plants 18 days after rub-inoculation with sap prepared from N. benthamiana leaves infected with purified virions of the tomato chocolate-spot-associated virus. These symptoms are indistinguishable from those observed in tomato plants in the field. f Necrosis symptoms developing in the basal portion of leaves of tomato plants (cv. El Senor) about three weeks after graft inoculation with shoots from N. benthamiana plants infected with purified virions of the tomato chocolate-spot-associated virus
Here, we describe and characterize a virus associated with the chocolate spot disease in Guatemala. The virus is sap- and graft-transmissible and induces disease symptoms in a number of solanaceous plants, including chocolate spot in tomatoes. This virus has icosahedral virions (~28–30 nm) and a genome composed of two single-stranded RNAs [(~7,500 and 5,100 nucleotides (nt)]. The chocolate-spot-associated virus is similar to recently described torradoviruses associated with necrosis diseases of tomato in Spain and Mexico, but sequence analysis indicated it is a member of a distinct species. Thus, the name tomato chocolate spot virus (ToCSV) is proposed.
Materials and methods
Virus transmission, propagation and host range
Samples of leaves and petioles with chocolate spot symptoms were collected from tomato plants during surveys conducted in Guatemala from 2003 to 2006. Tissues from these samples were squashed onto nylon and nitrocellulose membranes, which were returned to the University of California-Davis. Tissue squashes were excised and ground in 0.01 M potassium phosphate buffer, pH 7.2. The resulting sap was rub-inoculated onto leaves of N. benthamiana plants at the 3–5-leaf stage.
Leaf tissue from N. benthamiana plants that developed virus-like symptoms and tested negative for known tomato-infecting viruses was used for sap inoculation of additional N. benthamiana plants. In this way, the tomato chocolate-spot-associated virus was propagated in N. benthamiana plants, and these infected plants were used as the source of tissue for virion purification, host range determination and other experiments.
The partial host range of the chocolate-spot-associated virus was determined by rub-inoculating sap prepared from infected N. benthamiana plants onto leaves of seedlings of Capsicum annuum L. cv. Yolo Wonder, Chenopodium quinoa (Willd.), C. amaranticolor L., Cucurbita pepo cv. Small Sugar, Datura stramonium L., Solanum lycopersicum M. cv. Early Pak 7, Nicotiana benthamiana, N. tabacum L. cv. Havana, N. tabacum L. cv. Xanthi, N. glutinosa and Phaseolus vulgaris cv. Topcrop.
To further determine the nature of the symptoms induced by the chocolate-spot-associated virus in tomato, a series of cultivars were rub-inoculated with sap or grafted with shoots from N. benthamiana plants infected with purified virions. These included El Senor, Classy Lady, SUN6366, NUN126123, NUN2139, NUN6385, NUN6394, NUN12184, NUN12351, NUN13478, NUN2125 and NUN2115 (Nunhems); H5508 and H5608 (HeinzSeed); Roma (Tropica); PX002, DRI0309, Stevens and Celebrity (Seminis); Tsarine, Geneva 90, Sahel, Stevens, Silvana, Pascaline and Qualit 21 (Syngenta); HMX4801 (Harris Moran) and CXD243 (Campbells).
Virion purification and electron microscopy
Two methods were used for virion purification: (1) a minipurification method [19] and (2) a more extensive procedure designed for viruses with labile spherical virions. For the latter procedure, 60 g of leaf tissue from infected N. benthamiana plants (8–14 days post-inoculation) was ground in liquid nitrogen and homogenized in 70 ml of extraction buffer (0.5 M sodium citrate, pH 6.5; 5 mM EDTA; and 1 ml/L 2-mercaptoethanol). The homogenate was transferred into 250-ml polypropylene centrifuge tubes, which were centrifuged at 4,000×g for 5 min. The supernatant was removed and stirred with 1 volume of chloroform for 5 min, or until the suspension had been emulsified. This suspension was transferred into 30-ml polypropylene centrifuge tubes, which were centrifuged at 12,000×g for 10 min. The upper (aqueous) phase was removed and transferred to a 100-ml beaker, which was placed on ice. Polyethylene glycol (PEG) 8000 was added into this solution, with stirring, to a final concentration of 10%. The solution was adjusted to 0.1 M NaCl, stirred for 15 min, placed on ice for 30–40 min, and then centrifuged at 12,000×g for 20 min. The supernatant was discarded, and the pellet was suspended in 24 ml of suspension buffer (2.5 mM sodium tetraborate with 0.5 mM EDTA, pH 9.0). Triton X-100 was added to a final concentration of 2%, and the solution was stirred for 30 min. This solution was centrifuged at 19,000×g for 5 min, and the supernatant was transferred into 26.3-ml polycarbonate tubes for the 70 Ti rotor. These tubes were centrifuged at 311,080×g for 1.5 h. The pellet was retained and suspended in 15 ml of suspension buffer. This suspension was centrifuged at 12,000×g for 5 min, and the supernatant was transferred into polycarbonate ultracentrifugation tubes with 5 ml of suspension buffer in 25% (w/w) sucrose as a cushion. These tubes were placed in a 50.2 Ti rotor and centrifuged at 147,907×g for 1.5 h. The supernatant was discarded, and the pellet was resuspended in 1 ml of suspension buffer. This suspension was transferred into 1.5 ml Eppendorf tubes, which were centrifuged at 10,000×g for 5 min. The supernatant was then transferred onto a sucrose gradient [5–30% (w/w)] in SW 41 Ultra-Clear 13.2-ml tubes, which were centrifuged at 209,866×g for 1 h. The entire virion purification procedure was completed in one day to minimize virion degradation. The presence of virions was initially determined by analysis of fractions with SDS-PAGE and silver staining (Promega). Based on these results, fractions potentially having virions were collected with a 1-ml syringe by puncturing the tube and removing the appropriate part of the gradient. An aliquot of these fractions was stained with 2% uranyl acetate and examined for virus particles with a transmission electron microscope (JEOL JEM-1230, Japan).
Polyacrylamide gel electrophoresis
Proteins associated with purified virions were analyzed by SDS-PAGE in 12% denaturing gels [28]. Approximately 5 μl of the purified virion preparation was analyzed, and proteins were visualized by silver staining.
Protein identification by mass spectrometry
Aliquots of fractions having virus particles based on PAGE and electron microscope analyses were analyzed by SDS-PAGE. After silver staining, protein bands of interest were excised from the gel and sent for sequencing at the Proteomics Core Facility at UC Davis.
Nucleic acid extraction
RNA was extracted from purified virions, and total RNA was extracted from leaf tissues using a Qiagen RNeasy kit (Qiagen) according to the manufacturer’s instructions.
Double-stranded (ds) RNA was extracted from 15 g of leaf tissue from infected N. benthamiana plants. Enriched dsRNA was prepared by two cycles of chromatography on CF-11 cellulose columns, as described by Dodds and Bar-Joseph [9].
RNA concentrations were determined with a BioMate 3-spectrophotometer (Thermo Electron Corp. Madison, USA). The size and integrity of the putative viral RNAs were determined in 1% agarose gels with formaldehyde/formamide/HEPES buffer. The presence of dsRNA was determined by analysis of 10 μl of dsRNA preparations in 1% agarose gels with formaldehyde/formamide/HEPES buffer. Gels were stained with ethidium bromide and visualized with UV light. The dsRNA preparations were also examined by PAGE in 12% gels with 8 M urea, followed by silver staining [28].
cDNA synthesis, cloning and sequencing
Viral RNA from purified virions or dsRNA from infected N. benthamiana leaves were used as templates to synthesize double-stranded (ds) cDNA using the SuperScript Choice System (Invitrogen) according to the manufacturer’s instructions. First-strand cDNA was primed with either oligo(dT) or random hexamer primers (Invitrogen). After second-strand synthesis, ds-cDNA was cloned into the Zero Blunt TOPO PCR Vector (Invitrogen). TOP10 competent cells (Invitrogen) were transformed with the ligation mixture, and clones containing inserts ≥500 nt were sequenced.
Additional cDNA fragments, representing the remainder of the viral genome, were generated using total RNA extracts from infected N. benthamiana leaves, RT–PCR with SuperScriptII reverse transcriptase (Invitrogen), and PCR with iProof High-Fidelity DNA polymerase (Bio-Rad). Primers were designed from sequences of the cloned cDNA fragments and are shown in Table 1. PCR-amplified cDNA fragments were purified from agarose gels using a Qiaquick PCR Purification Kit (Qiagen) and either cloned into the Zero Blunt TOPO PCR Vector (Invitrogen) or directly sequenced at the UC Davis Sequencing Facility. Discrepancies between individual sequences were resolved by re-cloning and sequencing of multiple clones.
Table 1.
List of primers used in this study
| Primer | Sequence (5′–3′)a | Position (nt)b | Applications/virus detectionc |
|---|---|---|---|
| ToCSV1 | CGTGGCTTTATTTCCTCGAA | RNA1-3361 | RdRp gene specific |
| ToCSV2 | ATGGACTTAAGGATATCACAGC | RNA1-3461 | 5′RACE PCR |
| ToCSV3 | GGAGCTGAGCACTTCATGT | RNA1-3861 | 3′RACE PCR |
| ToCSV4 | TTGAGCTCAACGCGGTCC | RNA1-3965 | RdRp gene specific |
| ToCSV5 | ATCAGAAATATCCAAGTTACAAC | RNA2-636 | ORF1 specific |
| ToCSV6 | TTCCTGCGCCCTGTTACTT | RNA2-739 | 5′RACE PCR |
| ToCSV7 | GAGAGGACCAATGTTCTACT | RNA2-1147 | 3′RACE PCR |
| ToCSV8 | GGCACTCCTTGTATCTATCT | RNA2-1252 | ORF2 specific |
| DG19 | ATGKCTTTTTCMAAGATGTTCYCC | RNA1-140 | ToMarV, ToTV, ToANV |
| DG20 | CCATTKGGTCTACCAGGYACWG | RNA1-850 | ToMarV, ToTV, ToANV |
| DG21 | TCTYTATGTCATTYATTKSKCGTTT | RNA2-180 | ToMarV, ToTV, ToANV |
| DG22 | CCATARAGAAYCCCRTAKGAACCC | RNA2-880 | ToMarV, ToTV, ToANV |
| ToMarV36 | AAGCCATTGTAGAAGTTAGAAACT | RNA1-100 | ToMarV specific |
| ToMarV37 | TGGCGCAAGCCAGTATGTTAC | RNA1-710 | ToMarV specific |
| ToMarV38 | CTGCATATGGCATCACCAAGG | RNA2-1841 | ToMarV specific |
| ToMarV39 | GGAACCGCGGGAAATATGTTTG | RNA2-2880 | ToMarV specific |
| ToMarV40 | CAATGTCAAGATATCAAAGATCTC | RNA1-1500 | ToMarV specific |
| ToMarV41 | AAAGAATTATATATTGAGATTGCAACC | RNA1-1 | 5′ END specific |
| ToMarV42 | AACCCAGGAAAAGCCTAACTT | RNA1-500 | ToMarV specific |
| ToTV1 | TTTAAAAGAATAATTTTATACAATATT | RNA2-1 | ToTV specific |
| ToTV2 | GAGCATGGGCACCCGGA | RNA2-1502 | ToTV specific |
| ToTV3 | TTTTCATATGGGGCTGTACAA | RNA2-3645 | ToTV specific |
| ToTV4 | TAAAATACATATTCAAACTCACAC | RNA2-5390 | ToTV specific |
| ToTV5 | TTAAAAGAGTTATTTTGAGAATATA | RNA1-1 | ToTV specific |
| ToTV6 | GCCAAAGATGAGCGCTTGC | RNA1-4013 | ToTV specific |
| ToTV7 | AAAACCTCTGCTTAGAAATGTT | RNA1 -4768 | ToTV specific |
| ToTV8 | GTGAATAAGTCCGTAGACAAT | RNA1 -1120 | ToTV specific |
| ToANV1 | ACCAACATCTCAGTTGATGTC | RNA1-1 | ToANV specific |
| ToANV2 | CAAGGAGAGAGTTGTGGAG | RNA2-1 | ToANV specific |
| ToANV3 | ATTTTCTTTTCTTTTATTTC | RNAs-3′ | 3′ END specific |
| ToANV4 | CAGGATAGCTTGGTGAGCAT | RNA1-700 | ToANV specific |
| Poly(T) | AGCTGGATCCTTTTTTTTTTTTTTTTTV | General oligo(dT) primer |
ToCSV tomato chocolate spot virus, DG degenerate primer, ToTV tomato torrado virus, ToMarV tomato marchitez virus, ToANV tomato apex necrosis virus
aPrimer sequences were derived from sequences with the following GenBank accession numbers and references: ToCSV = GQ305131 (RNA1) and GQ305132 (RNA2); ToTV = DQ388879 (RNA1) and DQ388880 (RNA2) [32]; ToMarV = EF681764 (RNA1) and EF681765 (RNA2) [33]; and ToANV = EF063641 (RNA1) and EF63642 (RNA2) [31]. In the primer sequences, K = g/T; M = A/C; Y = C/T; W = A/T; S = C/g and V = A/C/g
bThe viral RNA (RNA1 or 2) and position of the nucleotide corresponding to the 3′ nucleotide of the primer
cThe application for which the primer was used or for which of the viruses the primer can be used for RT–PCR detection
To determine the sequences of the 5′ and 3′ ends of the viral RNAs, a Smart RACE cDNA Amplification Kit (Clontech) was used, and amplified fragments were sequenced by “walking” towards the 5′ and 3′ ends of the RNAs with virus-specific primers (Table 1). The templates used were viral ssRNA (1 μg) extracted from purified virions and dsRNA (90 ng) extracted from infected N. benthamiana tissues. The 5′ and 3′ ends of each of the two viral RNAs were determined at least three times with different ss- or dsRNA extracts.
Sequence analysis
Sequences were initially compared with those in GenBank using the BLAST program available at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/BLAST/). Overlapping sequences were assembled using the BioEdit program version 7.05 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). ORFs were identified using Vector NTI Advance 10 software (Invitrogen). Sequences were aligned using Clustal W [30]. Phylogenetic trees were constructed from the Clustal W-aligned sequences with MEGA 4 [18], using a neighbor-joining method to conduct the bootstrap analysis (1,000 replicates).
Results and discussion
Virus characterization
The appearance of chocolate spot disease in Guatemala in 1999 and subsequent yield losses to tomato production due to the disease, particularly in production areas at higher elevation, led to concerns that a new tomato-infecting virus had emerged or had been introduced. Further evidence for this came from the fact that samples of tomato leaves with chocolate spot symptoms tested negative for infection with known viruses that induce necrosis symptoms in tomato, including tomato spotted wilt virus (TSWV) and tobacco streak virus (TSV) as well as other known tomato-infecting viruses, including begomoviruses and potyviruses (data not shown).
When sap prepared from tomato leaf tissue with chocolate spot symptoms was rub-inoculated onto leaves of N. benthamiana plants, two types of disease symptoms developed: (1) systemic necrosis and (2) epinasty, leaf crumpling and yellowing. Immunostrip tests and indicator plant inoculations established that the systemic necrosis symptoms were caused by a tobamovirus, most likely tobacco mosaic virus (TMV) or tomato mosaic virus (ToMV). However, a tobamovirus was ruled out as the causal agent because chocolate spot symptoms developed in tobamovirus-resistant tomato cultivars, and tests of leaves with chocolate spot symptoms were frequently negative for tobamovirus infection.
In contrast, the epinasty, leaf crumpling and yellowing symptoms developed in some N. benthamiana plants inoculated with sap prepared from leaves with chocolate spot symptoms collected from tomato cultivars with and without tobamovirus resistance. Leaves from N. benthamiana plants with the epinasty, crumpling and yellowing symptoms also tested negative for infection with cucumber mosaic virus (CMV), potato virus Y, TMV/ToMV and TSWV. Furthermore, when shoots from N. benthamiana plants with these symptoms were grafted onto tomato plants, newly emerging shoots developed chocolate spot symptoms, including epinasty and necrotic spots on leaves, petioles and stems (data not shown). Together, these results suggested that this sap-transmissible virus was the causal agent of chocolate spot disease, and it was referred to as the chocolate-spot-associated virus. The virus was readily sap-transmitted from N. benthamiana to N. benthamiana, and it was therefore propagated in this host.
The host range of the chocolate-spot-associated virus was investigated by rub-inoculation of a series of indicator plants with sap prepared from leaves of symptomatic N. benthamiana plants. In addition to N. benthamiana, obvious disease symptoms developed in N. glutinosa and Datura stramonium. In N. glutinosa, symptoms included epinasty, mosaic and mottling of newly emerged leaves, followed by necrosis. In D. stramonium, leaves developed an interveinal yellow mottling and flecking. Inoculated N. tabacum plants showed very mild mottling. In these experiments, sap-inoculated tomato seedlings did not developed symptoms; however, we were subsequently able to sap-transmit the virus to some tomato cultivars (see below). No obvious symptoms were observed in sap-inoculated Chenopodium quinoa, Chenopodium amaranticolor, common bean or small sugar pumpkin plants. These results established that the chocolate-spot-associated virus is sap-transmissible and has a relatively narrow host range, limited to members of the Solanaceae.
To further characterize the chocolate-spot-associated virus, virus purification was performed. Initial attempts using a minipurification method [19] were unsuccessful, as examinations of preparations from N. benthamiana leaves infected with the chocolate-spot-associated virus with the electron microscope failed to reveal virus-like particles. In contrast, examinations of preparations from common bean leaves infected with bean common mosaic virus revealed the expected potyvirus virions (long flexuous rods). These results indicated that the minipurification technique was performed properly and that the virions of the chocolate-spot-associated virus may be labile or present in low concentration.
We next used a more extensive purification procedure designed for labile spherical virions, which involves density gradient centrifugation (see “Materials and methods”). Three putative virion-containing fractions were identified based upon the presence of candidate virus structural proteins as revealed by SDS-PAGE analysis (data not shown). Electron microscopy analysis revealed virus-like icosahedral particles with a diameter of ~28–30 nm in one of these fractions (Fig. 2a). Evidence that these were the virions of the chocolate-spot-associated virus came from the finding that N. benthamiana plants rub-inoculated with these purified virions developed epinasty, leaf crumpling and yellowing symptoms that were indistinguishable from those in N. benthamiana plants infected with the virus. The presence of the chocolate-spot-associated virus in N. benthamiana plants that had been infected with purified virions was subsequently confirmed by RT–PCR and sequencing (as described below).
Fig. 2.
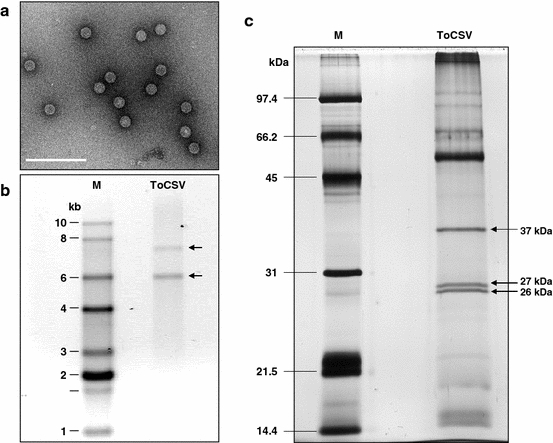
Characterization of virions, viral RNAs and structural proteins of the tomato chocolate-spot-associated virus (ToCSV). a Electron micrograph of negatively stained virions from a fraction recovered following sucrose density gradient centrifugation. Scale bar = 200 nm. b Ethidium-bromide-stained denaturing agarose gel showing RNAs extracted from purified virions shown in (a) compared with an RNA marker (M). Arrows indicate the position of the two RNAs associated with purified virions of the tomato chocolate-spot-associated virus. The marker used was the Invitrogen RNA Ladder, and sizes are indicated at the left in kilobases (kb). c Denaturing polyacrylamide gel electrophoresis (SDS-PAGE) of virions shown in (a) extracted from N. benthamiana plants infected with the tomato chocolate-spot-associated virus. Proteins were visualized by silver staining. Arrows indicate putative viral structural proteins, with estimated sizes shown at the right. Higher-molecular-weight protein bands are of host origin. Further efforts to purify the virions led to degradation of the structural proteins (data not shown). The marker (M) used was the Bio-Rad silver stain SDS-PAGE standards (low range), and protein sizes are indicated at the left in kilodaltons (kDa)
To confirm that the icosahedral virions of the chocolate-spot-associated virus were the causal agent of chocolate spot disease of tomato, sap prepared from symptomatic leaves or shoots from N. benthamiana plants that had been infected with purified virions was rub-inoculated or grafted onto tomato seedlings, respectively. Sap-inoculated tomato seedlings of cultivars El Senor, Classy Lady, NUN12351, NUN2115, NUN6385, NUN6394, NUN12184, NUN126123, H5508, H5608, PX002 and DRI0309 developed typical chocolate spot symptoms, including necrotic spots on basal portions of newly emerged leaves, epinasty and chlorosis (Fig. 1e). Similarly, tomato cultivars El Senor, NUN2115, NUN6385, NUN6394, NUN12184, NUN126123, H5508, H5608 and PX002 also developed typical chocolate spot symptoms when grafted with shoots from infected N. benthamiana plants (Fig. 1f). The presence of the chocolate-spot-associated virus in representative symptomatic leaves of sap-inoculated and grafted tomato plants was confirmed by RT–PCR analysis (data not shown). Initially, the overall rate of sap transmission was relatively low (1–3%); however; it was substantially increased (60–100%) by using young tomato seedlings held in the dark for 24 h and then maintained at a reduced temperature (22 vs. 26°C). This result is also consistent with chocolate spot disease being prevalent at higher elevations in Guatemala, where temperatures are lower. It is also interesting to note that a number of tomato cultivars did not develop symptoms following sap or graft inoculation. This suggests that some cultivars are resistant to infection by the chocolate-spot-associated virus. Together, these results indicated that the chocolate-spot-associated virus is the causal agent of chocolate spot disease of tomato.
Initial attempts to characterize the genome of the chocolate-spot-associated virus based on dsRNA properties were unsuccessful, as very low amounts of dsRNA (<100 ng) were recovered from leaves of infected N. benthamiana plants. Furthermore, dsRNAs were not observed in these dsRNA preparations in ethidium-bromide-stained agarose or silver-stained polyacrylamide gels (data not shown). In contrast, the expected-size dsRNA profiles were obtained from plants infected with TSWV, TMV, CMV and alfalfa mosaic virus (AMV), indicating that the dsRNA purification method was performed properly. This suggests that the dsRNA titer of the chocolate-spot-associated virus is low or that dsRNA accumulates transiently. However, when nucleic acids were extracted from purified virions and analyzed by denaturing agarose gel electrophoresis, two prominent ssRNA bands, having estimated sizes of 7.5 and 5.1 kb, were consistently detected (Fig. 2b).
Characterization of the structural proteins of the chocolate-spot-associated virus by SDS-PAGE analysis of purified virions revealed three putative viral structural proteins with estimated sizes of 26, 27 and 36 kDa, respectively (Fig. 2c). Here, it is important to note that other proteins were observed in these extracts, but comparisons with extracts prepared from uninfected N. benthamiana leaves indicated that these were of host origin (data not shown). Attempts to purify the chocolate-spot-associated virus further resulted in elimination of the host proteins but also altered the profile of the virion structural proteins, with the appearance of additional proteins. This suggested that the additional purification steps led to degradation of the virions (data not shown).
The morphology and dimensions of the virions, the number and size of the genomic RNAs and the number and approximate size of the structural proteins of the chocolate-spot-associated virus are similar to those of recently described picorna-like viruses associated with necrosis-associated diseases of tomato in Spain [torrado (meaning burned or roasted) disease] and in Mexico [marchitez (meaning wilted or withered) disease]. Torrado disease is caused by tomato torrado virus (ToTV) [32], whereas marchitez disease is caused by tomato apex necrosis virus (ToANV) [31] and tomato marchitez virus (ToMarV) [33], which appear to be isolates of a single species [29, 33]. The previously described ToANV [31], ToMarV [33] and ToTV [32] each have (1) spherical virions of ~28–30 nm in diameter; (2) two genomic RNAs with approximate sizes of 7 and 5 kb, 7.2 and 4.8 kb, and 7.7 and 5.3 kb; and (3) three structural proteins of 24, 28 and 38 kDa, 24, 26 and 35 kDa, or 23, 26 and 35 kDa, respectively. Based on these distinctive properties, these viruses have been placed in a new genus of picorna-like viruses, Torradovirus [29, 32]. Recently, the “plant picornavirales” ICTV study group reevaluated the taxonomy of this group of viruses and proposed a new family Secoviridae, which includes the genus Torradovirus [29]. Thus, the virions, genomic RNAs and the structural proteins of the chocolate-spot-associated virus are similar to those of these newly described torradoviruses; however, the sizes of the RNAs and the structural proteins were not identical in size.
The chocolate spot disease symptoms were similar to those of the torrado and marchitez diseases, and the properties of the chocolate-spot-associated virus were similar to those of the newly described torradoviruses. Thus, it was possible that one of these torradoviruses was the casual agent of chocolate spot disease. To assess this possibility, we used RNA extracts prepared from leaves with chocolate spot symptoms collected from various locations in Guatemala and from leaves of N. benthamiana and tomato plants infected with the chocolate-spot-associated virus in RT–PCR tests with ToTV- and ToANV-specific primer pairs (Table 1). As positive controls, we used RNA extracts prepared from leaves of ToTV- and ToANV-infected tomato plants (kindly provided by Craig Sandlin of Syngenta Co.). In these tests, the expected-size 0.7-kb fragments were amplified from RNA extracts prepared from ToTV-infected tissues with the ToTV6 and ToTV7 primer pair, and from ToANV-infected tissues with the ToANV1 and ToANV4 primer pair. In contrast, no fragments were amplified from RNA extracts prepared from tomato tissues with chocolate spot symptoms or leaves of N. benthamiana or tomato plants infected with the chocolate-spot-associated virus (data not shown). These results indicated that chocolate spot disease is not caused by ToANV or ToTV.
Cloning and sequence analysis of RNAs of the chocolate-spot-associated virus
Taken together, our results indicated that the chocolate-spot-associated virus may represent a new torradoviruses species. This was also consistent with our inability to identify a previously characterized virus as the cause of chocolate spot disease of tomato. Therefore, PCR primers were designed from conserved regions identified in the RNA1 and RNA2 sequences of ToANV, ToMarV and ToTV (Table 1 and data not shown) and used in RT–PCR tests with RNA extracted from purified virions and N. benthamiana leaf tissue infected with the chocolate-spot-associated virus. However, these “general” torradovirus RNA1 or RNA2 primers, individually paired with an oligo(dT) primer, failed to direct the amplification of cDNA from the chocolate-spot-associated virus RNAs or extracts from infected plants. This was consistent with previous results indicating that the chocolate-spot-associated virus was not one of the previously characterized torradoviruses.
Therefore, to characterize the RNAs of the chocolate-spot-associated virus, cDNA was generated from RNA extracted from purified virions and dsRNAs from infected N. benthamiana leaf tissues using random hexamers and an oligo(T) primer. This was accomplished via a two-step process in which single-stranded cDNA was initially synthesized, followed by a second reaction to generate the double-stranded cDNA. A total of 15 putative chocolate-spot-associated virus cDNA clones, all with inserts of ~500–600 bp, were obtained. Sequence analysis revealed one clone having identities of 75, 74 and 68% with RNA1 sequences of ToANV, ToMarV and ToTV, respectively, and in a region corresponding to the open reading frame (ORF) encoding the RNA-dependent RNA polymerase (RdRp). Another clone had identities of 78 and 72% with ORF1 of RNA2 of ToMarV and ToTV, respectively. The sequences of these clones were then used to design primers to facilitate the amplification and cloning of the remainder of the viral genome. A 5′and 3′-RACE sequence-walking strategy was used to generate and confirm sequences of the ends of both RNAs. Together, these sequences were used to assemble the complete sequences of the RNA1 and RNA2 of the chocolate-spot-associated virus.
RNA1
The RNA1 is 7,473 nt long [not including the poly (A) tail], with a single ORF (ORF1). The predicted ORF1 has an AUG at nt 139–141 and a UGA stop codon at nt 6,604–6,606, and encodes a predicted polyprotein of 2,155 amino acids (aa) with a molecular mass of 240 kDa (Fig. 3). A BLAST search with the complete nt sequence of the chocolate-spot-associated virus RNA1 revealed 74 and 69% identity with RNA1 sequences of ToMarV and ToTV, respectively, and 74% identity with the partial ToANV RNA1 sequence available in GenBank. The aa sequence of the predicted ORF1 polyprotein is 83% identical to those of ToMarV (EF681764) and ToANV (EF063641), and 64% identical to that of ToTV (DQ388879). These results are fully consistent with the chocolate-spot-associated virus representing a new torradovirus species.
Fig. 3.
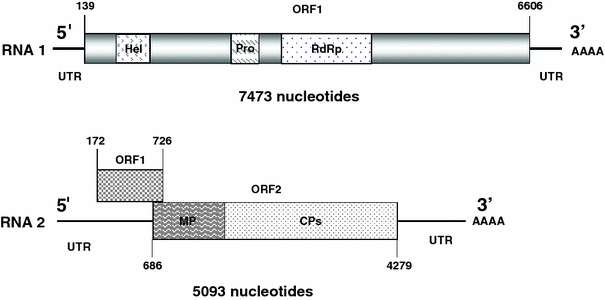
Genome organization of the tomato chocolate-spot-associated virus RNA1 and RNA2. Relative positions of the helicase (Hel), protease (Pro), and RNA-dependent RNA polymerase (RdRp) motifs in the polyprotein encoded by RNA1 are indicated. The relative locations of the putative movement protein (MP) and structural (capsid) proteins (CPs) in the RNA2-encoded polyprotein are indicated. Nucleotides corresponding to initiation (first nt of the AUG) and termination (last nt of the stop codon) codons are shown. ORF open reading frame, UTR untranslated region
Analysis of the predicted aa sequence of the ORF1 polyprotein revealed domains having similarity with helicase (Hel), protease (Pro) and RdRp motifs (Fig. 3). These motifs are associated with replication-associated proteins of many RNA viruses, and the module is a hallmark of members of the new family Secoviridae [4, 10, 11, 29]. Analysis of the helicase domain (aa 398–495) revealed the “A” GxxgxGK(S/T), “B” (D/E) (D/E) and “C” KgxxФxSФФФx(S/T)(S/T)N motifs, indicating that the chocolate-spot-associated virus helicase belongs to superfamily III, which includes members of the newly established family Secoviridae, including the torradoviruses [12, 22, 29].
The aa sequence of the helicase domain is 100% identical to those of ToMarV and ToANV, and 97% identical to that of ToTV. The viruses with the next highest levels of identity in this domain are the waikaviruses maize chlorotic dwarf virus (NP_619716) and rice tungro spherical virus (AAA66056), with 51 and 47% identity, respectively, and the fabavirus broad bean wilt virus 1 (BBWV-1; AY781171), with 32% identity. Analysis of the predicted RdRp motif of the chocolate-spot-associated virus revealed motifs I (KDE) through VII (FLSR) [16, 25], which spanned aa 1,307–1,555 of the predicted polyprotein. The aa sequence of this region of the chocolate-spot-associated RdRp was 93% identical to those of ToMarV and ToANV and 84% identical to that of ToTV. The next highest levels of identity were 40% with cherry rasp leaf virus (genus Cheravirus, AAW92113) and 31% for BBWV-2. Together, the helicase and RdRp aa sequence identities strongly support a close relationship among the chocolate-spot-associated virus, ToANV, ToMarV and ToTV and the placement of these viruses in the family Secoviridae.
A 3C cysteine protease (Pro 3C) domain was detected upstream of the RdRp region in the RNA1 polyprotein (aa 873–1,070; Fig. 3). It has been reported previously that 3C and 3C-like proteinases can have a cysteine or serine residue in the catalytic domain [20]. In the chocolate-spot-associated virus polyprotein, this domain has a chymotrypsin-like fold with a cysteine nucleophile (aa 1,044) instead of the more commonly found serine [3, 6, 23]. Sequence analysis of the ORF1 polyproteins of ToANV, ToMarV and ToTV also revealed the presence of this domain and the cysteine nucleophile.
RNA2
The RNA2 is 5,093 nt long [not including the poly (A) tail], with two partially overlapping ORFs (ORF 1 and 2; Fig. 3). A BLAST search with the complete sequence of the chocolate-spot-associated virus RNA2 sequence revealed 76, 75 and 70% identities with those of ToMarV, ToANV (partial sequence) and ToTV, respectively. The ORF1 has an AUG at nt 172–174 and a UAA stop codon at nt 724–726, and the ORF2 has an AUG at nt 686–688 and a UGA stop codon at nt 4,277–4,279. The RNA2 ORF1 encodes a predicted protein of 184 aa with molecular mass of 20 kDa, whereas ORF2 encodes a predicted protein of 1,197 aa with a molecular mass of 133 kDa.
The RNA2 ORF1 nt sequence is 70% identical with those of ToMarV and ToTV, whereas the aa sequence of the predicted protein is 70 and 62% identical, respectively. A BLAST search revealed no other sequences having substantial identities with the chocolate-spot-associated virus RNA2 ORF1. The RNA2 ORF2 nt sequence is 75% identical to those of ToMarV and ToANV and 71% identical to that of ToTV. The aa sequence of the predicted ORF2 protein is 83, 82 and 69% identical to those of ToMarV, ToANV and ToTV, respectively. The predicted polyprotein encoded by ORF2 has a genome organization similar to those of ToMarV and ToTV, with a putative movement protein (MP) at the N-terminus and multiple CPs at the C-terminus. Evidence that the N-terminal portion of the ORF2 polyprotein may be a torradovirus movement protein (MP) came from the identification of a MP consensus sequence motif (LxxPxL [24]), which is also found in the MPs of bromoviruses (e.g., brome mosaic virus) and cucumoviruses (e.g., CMV) [8]. In the chocolate-spot-associated virus, this motif was LRIPTL and was located at aa 263–268. However, conclusive evidence that this protein is the torradovirus MP will need to come from genetic or functional studies.
Based upon SDS-PAGE analysis of purified virions, the structural proteins of the chocolate-spot-associated virus are 37, 27 and 26 kDa. These three CPs are referred to as Vp37, Vp27 and Vp26 and are similar but not identical in size to the three structural proteins of ToANV (38, 28 and 24 kDa), ToMarV (35, 26 and 24 kDa) and ToTV (35, 26 and 23 kDa), respectively [31–33]. Comparison of the predicted aa sequence of the chocolate-spot-associated virus ORF2 polyprotein with sequences in GenBank revealed that aa 483–724 have 84, 83 and 71% identity with the large CP (35–38 kDa) of ToMarV, ToANV and ToTV, respectively, aa 729–965 have 90, 89 and 86% identity with the medium-sized CP (26–28 kDa) of ToANV, ToMarV and ToTV, respectively, and aa 981–1,197 have 88, 89 and 71% identity with the small CP (23–24 kDa) of ToANV, ToMarV and ToTV, respectively. Thus, these results were consistent with SDS-PAGE results indicating that the chocolate-spot-associated virus has three major CPs. The differences in the size of the torradovirus CPs may be real or reflect differences in the experimental methods used in these analyses. This cannot be resolved by identifying predicted cleavage sites in the polyprotein, as these sites are very diverse in plant picorna-like viruses, and their precise positions need to be determined experimentally [7].
To gain insight into the nature of the structural proteins, the 26-kDa protein (Fig. 2c) was purified and subjected to liquid chromatography-mass/mass spectrometry analysis. A set of 19 peptide sequences were obtained from the trypsin-digested protein and analyzed with Mascot (http://www.matrixscience.com), a search engine that uses mass spectrometry data to identify proteins from primary sequence databases. The results of this search gave 16 possible matches, with the highest score (81) corresponding to the ToMarV and to ToANV polyproteins. These peptides corresponded to sequences of the polyproteins encoded by the ToMarV and ToANV RNA2 ORF2; however, the corresponding sequences were aa 115–131, aa 855–861, aa 1,120–1,126 and aa 1,155–1,167. Thus, the mass/mass spectrometry analysis of the 26-kDa structural protein resulted in matches with all three torradovirus CPs (see above). This can be explained by degradation of structural proteins during purification/recovery processes or complexities in the processing of these structural proteins.
Untranslated regions (UTRs)
The 5′-UTRs of RNA1 and RNA2 are 138 and 171 nt, respectively. Other than the first 11 nt, which are identical, the sequences are highly divergent (45% identity). The 5′-UTR of the chocolate-spot-associated virus RNA1 is 84 and 83% identical to those of ToMarV and ToTV, respectively, whereas the 5′-UTR of RNA2 is 77% identical to that of the 5′-UTR of ToMarV-RNA2. Interestingly, the 5′-UTR of RNA2 did not have substantial sequence identity (47%) with that of ToTV.
The 3′-UTRs of RNA1 and RNA2 are 867 and 814 nt long, respectively, not including the poly (A) tail. The 3′-UTR sequence of the chocolate-spot-associated virus RNA1 is 75, 78 and 88% identical to those of ToANV, ToMarV and ToTV, respectively, whereas the 3′-UTR of RNA2 is 74, 76 and 85% identical to those of ToANV, ToMarV and ToTV, respectively.
The RNA1 and RNA2 3′-UTR sequences are 67% identical, with the highest identity (~75%) in the 3′ 360 nt. Sequence conservation in the 3′-UTRs of bipartite picorna-like viruses has been reported previously, e.g., 78% or greater identity for 3′-UTRs of nepovirus genomic RNAs, and this may indicate an important role in viral replication [15, 21]. The nepoviruses have also been placed into the family Secoviridae, and into the recently created subfamily Comovirinae [29].
Our results established that the chocolate-spot-associated virus shares properties (e.g., host range, virion morphology, structural proteins, genome organization and sequence) with ToANV, ToMarV and ToTV. Furthermore, all of these viruses induce necrosis-associated diseases of tomato. However, there are also a number of differences between the chocolate-spot-associated virus and the previously characterized torradoviruses. For example, ToANV and ToMarV induce necrotic local lesions in Chenopodium quinoa plants, whereas the chocolate-spot-associated virus and ToTV do not induce symptoms in this host. There are also differences among these viruses in terms of the size of the genomic RNAs and the structural proteins. Perhaps most importantly, the RNA1 and 2 sequences of the chocolate-spot-associated virus were only ~80% identical with those of ToANV, ToMarV and ToTV. Thus, our results indicate that the chocolate-spot-associated virus is a member of a distinct torradovirus species, and the name tomato chocolate spot virus (ToCSV) is proposed.
Taxonomic position of ToCSV
Previous studies have shown that the RdRp is more informative for phylogenetic analyses of the positive-strand RNA viruses than is the CP [16, 17]. Indeed, this region has been used recently as the basis for the establishment of the order Picornavirales, which includes viruses in the family Picornaviridae and the newly established family Secoviridae [22, 29]. Thus, a phylogenetic analysis was performed with the aa sequence of the polymerase region of the ToCSV RNA1 polyprotein (between the CG protease motif [4] and the GDD RdRp active site [5]) to further assess the relationships among ToCSV, ToANV, ToMarV, ToTV and other picorna-like viruses, including those in the families Secoviridae and Picornaviridae (including viruses in the subfamily Comovirinae; and the genera Torradovirus, Sequivirus, Waikavirus, Sadwavirus, and Cheravirus). ToCSV was placed in a distinct clade with ToANV, ToMarV and ToTV (Fig. 4a). A similar result was obtained in a phylogenetic analysis conducted with the helicase region (between the motifs A and C; aa 397–494 [12]) (Fig. 4b). Together, these results clearly indicate that ToCSV, ToANV, ToMarV and ToTV are a closely related group of viruses, which belong together in the newly established genus Torradovirus. Furthermore, the torradoviruses have an RNA2 with two ORFs, which distinguishes them from other picorna-like viruses. The phylogenetic analyses also indicated that ToCSV, ToANV and ToMarV are more closely related to each other than to ToTV, likely reflecting the different geographical origin of these viruses, i.e., ToCSV, ToANV and ToMarV have New World origins, whereas ToTV has an Old World origin.
Fig. 4.
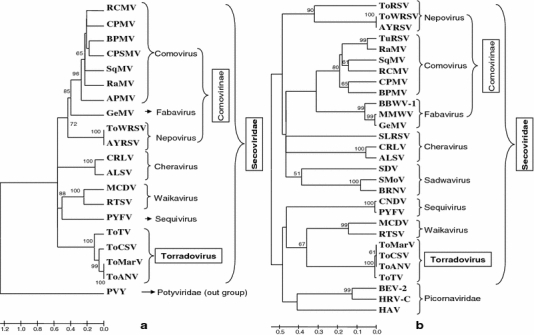
Phylogenetic analysis of tomato chocolate spot virus (ToCSV) and other picorna-like viruses based on alignments of a the region of the polyprotein between the protease CG motif and the GDD RdRp motif (aa 1044–1501) and b the helicase region between the motifs A and C (aa 397–494) of the RNA1-encoded polyprotein. The families and genera of the viruses are indicated on the right and the new genus, subfamily and family are shown in boxes. Sequences of other viruses used in these analyses are (virus abbreviations and accession numbers in parentheses): Andean potato mottle virus (APMV; Q02941), apple latent spherical virus (ALSV; NP_620568), artichoke yellow ringspot virus (AYRSV; CAJ33467), broad bean wilt virus-1 (BBWV-1; AAX12375), bovine enterovirus type 2 (BEV-2; AAS86317), bean pod mottle virus (BPMV; AF394608), black raspberry necrosis virus (BRNV; YP_654555), carrot necrotic dieback virus (CNDV; ACJ04421), cherry rasp leaf virus (CRLV; AAW92113), cowpea mosaic virus (CPMV; NP_734057), gentian mosaic virus (GeMV; BAD9900), hepatitis A virus (HAV; NP_041008), human rhinovirus C (HRV-C; YP_001552438), maize chlorotic dwarf virus (MCDV; AAV86083), mikania micrantha wilt virus (MMWV; ABX83032), parsnip yellow fleck virus (PYFV; NP_734447), radish mosaic virus (RaMV; YP_001911126), red clover mottle virus (RCMV; NP_734030), rice tungro spherical virus (RTSV; Q91PP5), satsuma dwarf virus (SDV; NP_734025), squash mosaic virus (SqMV; NP_734012), strawberry mottle virus (SMoV; NP_599086), strawberry latent ringspot virus (SLRSV; YP_227367), tomato ringspot virus (ToRSV; AAD50649), tomato white ringspot virus (ToWRSV; ABM65095), turnip ringspot virus (TuRSV; ABS90367), tomato chocolate spot virus (ToCSV; YP_003097229), tomato marchitez virus (ToMarV; YP_001976147), tomato torrado virus (ToTV; YP_001039627), and tomato apex necrosis virus (ToANV; ABK33525). Potato virus Y (PVY; ABA28320) was used as an outgroup sequence in the analyses. The numbers at each node are the bootstrap values (1000 replicates), and values >50% are shown. Scale bar and horizontal lines are in proportion to the number of amino acid differences between branch nodes
Development of a primer pair for detection of ToCSV by RT–PCR
We next performed alignments among the sequences of the RNA1 and RNA2 sequences of ToCSV, ToANV, ToMarV and ToTV to identify divergent sequences that could be used to design a specific primer pair for RT–PCR detection of ToCSV. A primer pair (ToCSV1 and ToCSV4; Table 1) was designed from a divergent region in the RNA1 sequence. This primer pair directed the amplification of the expected-size 600-bp fragment from RNA extracts prepared from tomato and N. benthamiana tissues infected with ToCSV, but not from RNA extracts from tomato tissue infected with ToANV or ToTV (Fig. 5). Thus, this primer pair can be used in RT–PCR assays to detect ToCSV.
Fig. 5.
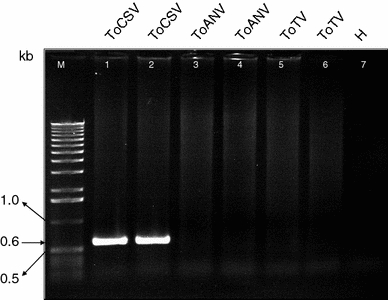
Specific detection of tomato chocolate spot virus (ToCSV) by RT–PCR with primer pair ToCSV1 and ToCSV4. Total RNA extracts were prepared from leaf tissue of tomato plants infected with ToCSV, tomato apex necrosis virus (ToANV) and tomato torrado virus (ToTV). The expected-size ~600-bp fragment was amplified from extracts from ToCSV-infected leaves (lanes 1 and 2), whereas no fragment was amplified from extracts from leaves infected with ToANV (lanes 3 and 4) or ToTV (lanes 5 and 6) or an extract prepared from leaves from an uninfected tomato plant (H; lane 7). M marker (1 kb DNA Ladder, Invitrogen)
The origin of these new tomato-infecting picorna-like viruses is not known. ToCSV represents the third such virus (presuming that ToANV and ToMarV are isolates of the same species) described within the past 4 years, indicating that these are a group of emerging viruses [27, 29, 31–33]. The recent emergence of these viruses and the fact that they are genetically distinct (~20% divergence in nt sequence) suggest that they do not represent a single species that has been disseminated via seed or propagative parts (e.g., tomato transplants), such as the tomato-infecting potexvirus, pepino mosaic virus [35]. It appears more likely that these viruses have emerged locally, perhaps by their introduction from unidentified reservoir hosts by an insect vector, possibly the greenhouse whitefly (Trialeurodes vaporariorum) [26]. A role for the greenhouse whitefly in the emergence and spread of chocolate spot disease in Guatemala is fully consistent with the association of these insects with disease outbreaks in the field. The emergence of the torradoviruses may also have been facilitated by the introduction and widespread cultivation of certain highly susceptible hybrid tomato varieties (J. Kao, personal communication). Secondary spread may then occur via movement of the insect vector or infected plant material; this may explain the recent reports of ToTV in the Canary Islands, Panama, Poland, Hungary, France and Australia [1, 2, 13, 14, 26, 34].
In this paper, we describe ToCSV, a member of a new torradovirus species that causes chocolate spot, a necrosis-associated disease of tomato in Guatemala. ToCSV is related to, but distinct from, ToANV and ToMarV from Mexico [31, 33] and ToTV from Spain [32]. Thus, ToCSV is another member of the genus Torradovirus in the family Secoviridae, a new family of picorna-like viruses that was recently approved by the ICTV [29, 32].
Acknowledgments
This project was funded by grants from the Ministry of Agriculture of Guatemala and PROYECTO AGROCYT No. 13-2004 funded by CONCYT and the Integrated Pest Management Collaborative Research Support Program (IPM-CRSP). This project was made possible by the United States Agency for International Development and generous support of the American people through USAID Cooperative Agreement No. EPPA-00-04-00016-00. We thank Hui-Ting Chou and Po-Lin Chiu for their assistance in transmission electron microscopy in the laboratory of Henning Stahlberg in the Department of Biophysics at UC Davis.
Conflict of interest statement
None.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The authors O. Batuman, Y.-W. Kuo contributed equally to this work.
Nucleotide sequence data are available in the GenBank under accession numbers GQ305131 (RNA1) and GQ305132 (RNA2).
An erratum to this article can be found at http://dx.doi.org/10.1007/s00705-010-0738-5
References
- 1.Alfaro-Fernández A, Córdoba-Sellés C, Cebrián MC, Sánchez-Navarro JA, Espino A, Martín R, Jordá C. First report of tomato torrado virus in tomato in the Canary Islands, Spain. Plant Dis. 2007;91:1060. doi: 10.1094/PDIS-91-8-1060B. [DOI] [PubMed] [Google Scholar]
- 2.Alfaro-Fernández A, Bese G, Córdoba-Sellés C, Cebrián MC, Herrera-Vásquez JA, Forray A, Jordá C. First report of tomato torrado virus infecting tomato in Hungary. Plant Dis. 2009;93:554. doi: 10.1094/PDIS-93-5-0554C. [DOI] [PubMed] [Google Scholar]
- 3.Allaire M, Chernaia MM, Malcolm BA, James MN. Picornaviral 3C cysteine proteinases have a fold similar to chymotrypsin-like serine proteinases. Nature. 1994;369:72–76. doi: 10.1038/369072a0. [DOI] [PubMed] [Google Scholar]
- 4.Argos P, Kamer G, Nicklin MJ, Wimmer E. Similarity in gene organization and homology between proteins of animal picornaviruses and a plant comovirus suggest common ancestry of these virus families. Nucleic Acids Res. 1984;12:7251–7267. doi: 10.1093/nar/12.18.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bazan JF, Fletterick RJ. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc Natl Acad Sci USA. 1988;85:7872–7876. doi: 10.1073/pnas.85.21.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blom N, Hansen J, Blaas D, Brunak S (1996) Cleavage site analysis in picornaviral polyproteins: Discovering cellular targets by neural networks. Protein Sci 5: 2203–2216 [DOI] [PMC free article] [PubMed]
- 8.Canto T, Prior DA, Hellwald KH, Oparka KJ, Palukaitis P. Characterization of cucumber mosaic virus, IV: movement protein and coat protein are both essential for cell-to-cell movement of cucumber mosaic virus. Virology. 1997;237:237–248. doi: 10.1006/viro.1997.8804. [DOI] [PubMed] [Google Scholar]
- 9.Dodds JA, Bar-Joseph M. Double-stranded RNA from plants infected with closteroviruses. Phytopathology. 1983;73:419–423. doi: 10.1094/Phyto-73-419. [DOI] [Google Scholar]
- 10.Franssen H, Leunissen J, Goldbach R, Lomonossoff G, Zimmern D. Homologous sequences in non-structural proteins from cowpea mosaic virus and picornaviruses. EMBO J. 1984;3:855–861. doi: 10.1002/j.1460-2075.1984.tb01896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldbach R. Genome similarities between plant and animal RNA viruses. Microbiol Sci. 1987;4:197–202. [PubMed] [Google Scholar]
- 12.Gorbalenya AE, Koonin EV, Wolf YI. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-I. [DOI] [PubMed] [Google Scholar]
- 13.Herrera-Vasquez JA, Alfaro-Fernández A, Cordoba-Selles MC, Cebrian MC, Font MIJC. First report of tomato torrado virus infecting tomato in single and mixed infections with cucumber mosaic virus in Panama. Plant Dis. 2009;93:198. doi: 10.1094/PDIS-93-2-0198A. [DOI] [PubMed] [Google Scholar]
- 14.International Phytosanitary Portal (IPPC F, Rome) (2008) Detection of tomato torrado virus in South Australia. (report AU-14/1 of 2008-10-10). https://www.ippc.int
- 15.Jonczyk M, Le Gall O, Palucha A, Borodynko N, Pospieszny H. Cloning and sequencing of full-length cDNAs of RNA1 and RNA2 of a tomato black ring virus isolate from Poland. Arch Virol. 2004;149:799–807. doi: 10.1007/s00705-003-0261-z. [DOI] [PubMed] [Google Scholar]
- 16.Koonin EV. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 17.Koonin EV, Dolja VV. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane LC. A general method for detecting plant viruses. In: Maramorosch K, editor. Plant diseases of viral, viroid, mycoplasma and uncertain origin. New Delhi: Oxford & IBH Publishing; 1992. pp. 3–17. [Google Scholar]
- 20.Lawson MA, Semler BL. Poliovirus thiol proteinase 3C can utilize a serine nucleophile within the putative catalytic triad. Proc Natl Acad Sci USA. 1991;88:9919–9923. doi: 10.1073/pnas.88.22.9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Gall O, Candresse T, Dunez J. Transfer of the 3′ non-translated region of grapevine chrome mosaic virus RNA-1 by recombination to tomato black ring virus RNA-2 in pseudorecombinant isolates. J Gen Virol. 1995;76:1285–1289. doi: 10.1099/0022-1317-76-5-1285. [DOI] [PubMed] [Google Scholar]
- 22.Le Gall O, Christian P, Fauquet CM, King AM, Knowles NJ, Nakashima N, Stanway G, Gorbalenya AE. Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch Virol. 2008;153:715–727. doi: 10.1007/s00705-008-0041-x. [DOI] [PubMed] [Google Scholar]
- 23.Matthews DA, Smith WW, Ferre RA, Condon B, Budahazi G, Sisson W, Villafranca JE, Janson CA, McElroy HE, Gribskov CL, et al. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 24.Mushegian AR. The putative movement domain encoded by nepovirus RNA-2 is conserved in all sequenced nepoviruses. Arch Virol. 1994;135:437–441. doi: 10.1007/BF01310028. [DOI] [PubMed] [Google Scholar]
- 25.O’Reilly EK, Kao CC. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 26.Pospieszny H, Borodynko N, Obrepalska-Steplowska A, Hasiow B. The first report of tomato torrado virus in Poland. Plant Dis. 2007;91:1364. doi: 10.1094/PDIS-91-10-1364A. [DOI] [PubMed] [Google Scholar]
- 27.Rojas MR, Gilbertson RL (2008) Emerging plant viruses: a diversity of mechanisms and opportunities. In: Roossinck MJ (ed) Plant virus evolution, Springer-Verlag, Berlin, pp 27–51
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanfacon H, Wellink J, Le Gall O, Karasev A, van der Vlugt R, Wetzel T. Secoviridae: a proposed family of plant viruses within the order Picornavirales that combines the families Sequiviridae and Comoviridae, the unassigned genera Cheravirus and Sadwavirus, and the proposed genus Torradovirus. Arch Virol. 2009;154:899–907. doi: 10.1007/s00705-009-0367-z. [DOI] [PubMed] [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turina M, Ricker MD, Lenzi R, Masenga V, Ciuffo M. A severe disease of tomato in the Culiacan area (Sinaloa, Mexico) is caused by a new picorna-like viral species. Plant Dis. 2007;91:932–941. doi: 10.1094/PDIS-91-8-0932. [DOI] [PubMed] [Google Scholar]
- 32.Verbeek M, Dullemans AM, van den Heuvel JF, Maris PC, van der Vlugt RA. Identification and characterisation of tomato torrado virus, a new plant picorna-like virus from tomato. Arch Virol. 2007;152:881–890. doi: 10.1007/s00705-006-0917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verbeek M, Dullemans AM, van den Heuvel JF, Maris PC, van der Vlugt RA. Tomato marchitez virus, a new plant picorna-like virus from tomato related to tomato torrado virus. Arch Virol. 2008;153:127–134. doi: 10.1007/s00705-007-1076-0. [DOI] [PubMed] [Google Scholar]
- 34.Verdin E, Gognalons P, Wipf-Scheibel C, Bornard I, Ridray G, Schoen L, Lecoq H. First report of tomato torrado virus in tomato crops in France. Plant Dis. 2009;93:1352. doi: 10.1094/PDIS-93-12-1352C. [DOI] [PubMed] [Google Scholar]
- 35.Verhoeven JTJ, van der Vlugt RAA, Roenhorst JW. High similarity between tomato isolates of pepino mosaic virus suggests a common origin. Eur J Plant Pathol. 2003;109:419–425. doi: 10.1023/A:1024261121468. [DOI] [Google Scholar]


