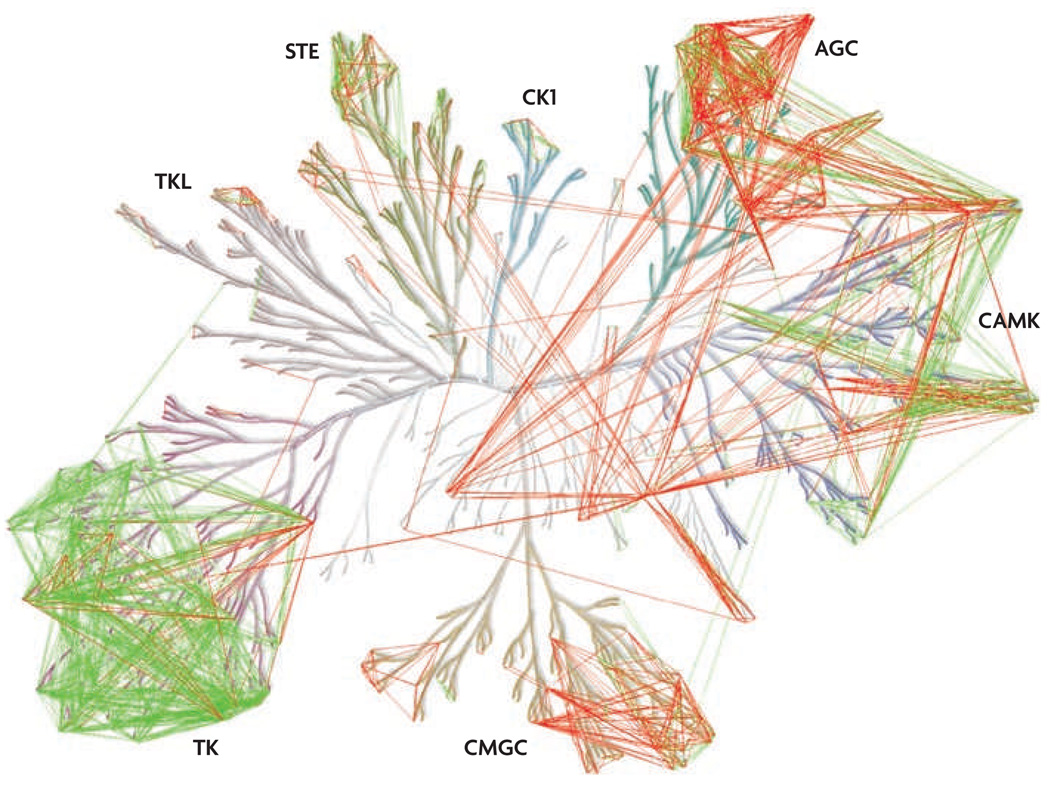
Figure 5. Polypharmacology in the protein kinome.
Pairs of kinases were associated based on kinase selectivity data for a broad range of inhibitor scaffolds93 and the sequence conservation between aligned sequences of an expanded ATP binding site. Pairs of kinases that were potently inhibited by a common inhibitor (common dissociation constant (KD)<10 nM) (green lines) were used to determine a sequence similarity cut-off to predict pairs of kinases that can be inhibited by a common inhibitor (red lines). A much higher sequence similarity in the binding pockets was observed for those pairs of kinases with a common potent inhibitor compared with pairs of kinases with a common but less potent inhibitor or no common inhibitor at all. The level of sequence conservation necessary for the predictions was determined based on the sequence conservation distribution of the experimentally determined pairs. The predicted pairs of kinases (red lines) represent potential target combinations that may be more easily accessible with new kinase inhibitors. The image is shown in more detail in supplementary information s1 (figure). AGC, kinases from the protein kinase A, protein kinase G and protein kinase C families; CAMK, calcium/calmodulin-dependent protein kinases; CK1, casein kinase 1; CMGC, kinases from the cyclin-dependent kinase, MAP kinase, glycogen synthase kinase and casein kinase II families; STE, homologues of yeast sterile 7, sterile 11 and sterile 20 kinases; TK, tyrosine kinases; TKL, tyrosine kinase-like kinases.