Abstract
The cerebellins are a family of four secreted proteins, two of which, Cbln1 and Cbln3, play an important role in the formation and maintenance of parallel fiber–Purkinje cell synapses. We have identified the chicken homologue of Cbln2 and, through the use of in situ hybridization, shown that it is expressed by specific subsets of neurons in the dorsal root ganglia (DRGs) and spinal cord starting shortly after those neurons are generated. In the developing spinal cord, Cbln2 is highly expressed by dI1, dI3, dI5, and dILB dorsal interneurons, to a lesser extent by dI2, dI4, dI6, and dILA dorsal interneurons, but not by ventral (v0 – v3) interneurons. After the spinal cord has matured and neurons have migrated to their final destinations, Cbln2 is abundant in the dorsal horn. In the DRGs, Cbln2 is expressed by TrkB+ and TrkC+ sensory neurons, but not by TrkA+ sensory neurons. Interestingly, regions of the spinal cord where TrkB+ and TrkC+ afferents terminate (i. e. laminae II, III, IV, and VI), exhibit the highest levels of Cbln2 expression. Cbln2 is also expressed by preganglionic sympathetic neurons and their targets in the sympathetic chain ganglia. Thus, the results show that Cbln2 is frequently expressed by synaptically connected neuronal populations. This, in turn, raises the possibility that if Cbln2, like Cbln1, plays a role in the formation and maintenance of synapses, it may somehow mediate bi-directional communication between discrete populations of neurons and their appropriate neuronal targets.
Keywords: Dorsal root ganglia, afferent projections, spinal cord, synaptic connections
INTRODUCTION
Within any given region of the nervous system, numerous distinct neuron types are present, each having a unique pattern of connectivity. For a properly functioning nervous system to form, it is absolutely essential that neurons project along the correct pathways and establish connections with the appropriate target cells during development. Despite the attention devoted to these issues for the last 2–3 decades, much remains unknown about how the axons of specific populations of neurons project through a complex environment and synapse on appropriate target neurons in a distant location.
We have focused on trying to understand how sensory axons choose the correct pathways to grow along, using the developing chick hindlimb as a model system. We have demonstrated that sensory axons projecting along different nerves are different from one another, responding differentially to an array of cues in their environment (Honig et al., 1998). Although some of those cues have been identified (Guan and Condic, 2003; Honig et al., 2005, Munoz et al., 2005), a full understanding of sensory axon pathfinding has been elusive. Accordingly, we have sought to identify differences in gene expression among different types of sensory neurons and attempted to do so by using a suppressive subtractive hybridization approach. One such screening yielded a sequence that we subsequently identified as part of the chicken homologue of cerebellin2 (Cbln2). In situ hybridization showed that Cbln2 is expressed by specific populations of neurons both in the dorsal root ganglia (DRGs) and in the spinal cord. Moreover, the neuronal populations expressing Cbln2 very frequently are synaptically connected.
The cerebellin family of genes consists of four members, termed Cbln1–4 (Bao et al., 2005). Cblns are secreted glycoproteins characterized by a conserved C-terminal globular C1q domain that mediates the formation of trimers (Urade et al., 1991; Pang et al., 2000) and an N-terminal cysteine motif that mediates the assembly of the trimers into higher order complexes (Bao et al, 2005). Cbln1, the first member identified (Urade et al, 1991), is the precursor of a hexadecapeptide that is enriched in the cerebellum and hence was named cerebellin (Slemmon et al., 1984). Granule cells synthesize Cbln1 (Pang et al., 2000) and release it from their parallel fiber (PF) terminals onto the dendritic spines of Purkinje cells (Miura et al., 2009), from which it is subsequently internalized (Wei et al., 2009). In the absence of Cbln1, there are multiple deficits in the structure and function of PF–Purkinje cell synapses (Hirai et al., 2005). Although the density of dendritic spines on Purkinje cells is not altered, the vast majority of them (~80%) lack presynaptic contacts with parallel fibers. Further, for the 20% of spines with presynaptic terminals, the postsynaptic densities are enlarged and the synapses show impaired neurotransmission and lack long-term depression. Thus, Cbln1 plays an important role in regulating synapse development, maintenance, and plasticity. Cbln3 forms heteromeric complexes with Cbln1 both in vitro and in vivo (Pang et al., 2000; Bao et al., 2006) and is associated with Cbln1 at PF-Purkinje cell synapses (Miura et al., 2009). However, Cbln3-null mice have no observable synaptic deficits, although the level of Cbln1 protein in the cerebellum is markedly increased (Bao et al., 2006), and so, the biological role of Cbln3 remains enigmatic.
Whereas Cbln1 is important for the normal development of PF–Purkinje cell synapses and for the maintenance of these synapses in the mature cerebellum, virtually nothing is known about the function of Cblns 2 and 4. Cbln2 is expressed at very low levels in the adult cerebellum (J. Morgan, personal communication) and so its function cannot be readily studied in the context of PF–Purkinje cell synapses. However, Cbln2 is abundant in many other parts of the adult and developing brain (Wada and Ohtani, 1991; Miura et al., 2006; Allen Brain Atlas; Reiner, Yang, Honig unpublished observations). Here we describe the spatial and temporal pattern of expression for Cbln2 in the DRGs and spinal cord during development in the chick.
MATERIALS AND METHODS
Identification of chicken Cbln2
A suppressive subtractive hybridization screening, which we had carried out to detect genes differentially expressed between sensory neurons projecting along different peripheral nerves, yielded an 180bp long sequence (BB180 clone). In situ hybridization showed expression in a subset of DRG neurons and in restricted regions of the spinal cord. This sequence was mapped to chicken chromosome 2 using the BLAT program (http://genome.ucsc.edu/cgi-bin/hgBlat). We then searched the NCBI EST database and found 15 EST clones clustered in the same genomic region and having overlapping sequences (BU392897, BU464164, BU279212, BU365548, BU280682, BU441998, BU383572, BU265266, BU464152, BU353757, CO419709, BU457636, CD216378, CN225199, and BM440526). We next assembled these clones into one 2.7kb-long contiguous sequence or “contig” (Genbank accession number GU189513), using the Vector NTI program (Invitrogen). As described in the Results section, we believe that this assembled contig represents the chicken homologue of Cbln2.
It should be noted that until the time of this report, chicken Cbln2 had been predicted in the NCBI database (Genbank accession number XM_419105). XM_419105 lacks ~500nt of the 5’ sequence we report here and therefore, does not contain the true translational initiation ATG codon. It would encode a protein 100aa shorter than that predicted by our contig.
Genbank search for other chicken Cblns
The only member of the Cbln family that has been correctly identified in the NCBI chicken database is Cbln4 (Genbank accession number NP_001072955).
The chicken Cbln1 sequence was obtained as Genbank accession number XM_001233212 and the alternative splicing variant XM_001233228, which are based on the genomic sequence. We found, by aligning 13 ESTs (BU463415, BU379146, BU318249, BU223879, BX272472, BU141765, BU140889, BU342081, BU142568, CV040776, BU141287, AL587661, and BX272472), that the genomic sequence contains an error whereby one nucleotide (adenine) is missing at position nt509. This one-nucleotide deletion results in a frame-shifting change in the coding region. The corrected mRNA sequence for XM_001233212 encodes a protein that is highly homologous to the mammalian Cbln1, especially at the C1q domain, and thus represents the bona fide chicken Cbln1 (Fig. 2B). The second clone, XM_001233228, has a 46nt deletion roughly in the middle of the region coding for the C1q domain, that causes an additional frame-shift and thus encodes a different protein sequence.
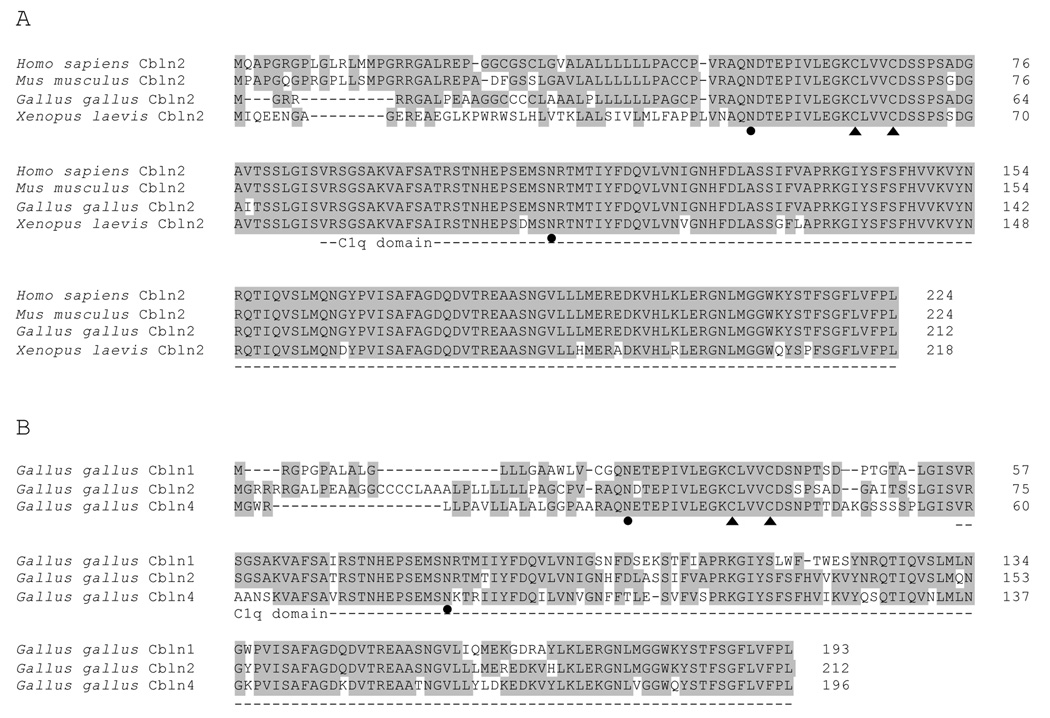
Figure 2. Cbln2 from Gallus gallus is highly homologous to Cbln2 from other species and to Cbln1 and Cbln4 from Gallus gallus.
A) Cbln2 from human (Genbank accession number NP_872317), mouse (Genbank accession number NP_766221), chicken (translated from Genbank accession number GU189513), and frog (Genbank accession number NP_001080811) are highly conserved, especially in the C1q domain (dotted line). B) Cbln2 from Gallus gallus shares homology with other cerebellin family members, Cbln1 (translated from Genbank accession number XM_001233212, modified as described in Materials and Methods) and Cbln4 (Genbank accession number NP_001072955). Cbln3 is believed to be absent in Gallus gallus. Conserved amino acid residues are shown in gray. The conserved cysteine residues for oligomerization are indicated by filled triangles. The conserved asparagines residues for N-glycosylation are indicated by filled circles.
To identify the chicken Cbln3 homologue, we used the consensus sequence of Cbln3 from human (NP_001034860), mouse (NP_062794), horse (XP_001488856), and dog (XP_547748) to BLAST the Genbank protein database and EST database for chicken. We found homologous sequences, but they all belonged to chicken Cbln4, Cbln2, or Cbln1, and no sequences corresponding to Cbln3 could be identified. Thus, the chicken genome does not appear to have a Cbln3 homologue.
Riboprobes for in situ hybridization
Antisense riboprobes used for in situ hybridization were prepared as follows. Total mRNA was isolated from embryonic chick DRGs using the RNeasy mini kit (Qiagen). Reverse transcription was performed with random hexamer primers and the SuperScript III First-Strand Synthesis System (Invitrogen). Specific target sequences (see below) were PCR amplified from the cDNA, cloned into PCRII-TOPO vector (Invitrogen), and the sequences later verified by the University of Tennessee Molecular Resources Center. The probes were transcribed using T7 or SP6 RNA polymerase with an NTP mixture containing either digoxigenin-11-UTP (DIG; Roche Applied Science) or dinitrophenol-11-UTP (DNP; Perkin Elmer). Riboprobe concentrations were estimated by denaturing agarose gel electrophoresis and for DIG-labeled probes, quantified using a dot blotting assay (Roche Applied Science), and were used at 100ng/ml.
The Cbln2 probe was directed to nucleotides 386–2218 (GenBank accession number GU189513), using CAGAACGACACGGAGCCCAT as the forward primer, and CCAGTACCGATTGCTTTGCAC as the reverse primer (Fig. 1). The 1800nt-long probe thus includes 500nt from the coding region (exons 2–5) and 1300nt from the 3’UTR. The 500nt portion from the coding region shares 78% and 71% identity with that of chicken Cbln1 and Cbln4, respectively, whereas the 3’UTR is unique to chicken Cbln2. Given that we used stringent hybridization conditions (calculated to distinguish sequences of 80% homology) and because the labeling pattern revealed by this probe was identical to those obtained with two other probes corresponding to parts of the 3’UTR (the original BB180 probe, corresponding to nt1953–2213, and another probe we made, corresponding to nt1453–2640), the pattern of expression we observed must accurately represent that of Cbln2.
Figure 1. Cbln2 from Gallus gallus.
The open reading frame that encodes chicken Cbln2 is underlined, and the predicted amino acid sequence is shown. The forward and reverse primers used for PCR are indicated. The inverted triangles mark the intron-exon splicing junctions predicted by comparing the mRNA sequence and the genomic sequence. Note that the junction between exon 2 and exon 3 has not been determined because the genomic sequence in this region is incomplete.
The TrkA probe was directed to nucleotides 297–1190 (GenBank accession number NM_205378), which codes for much of the extracellular domain of chicken TrkA. The forward primer was GATGCTTTCCAGGACAACCACA, and the reverse primer was GTTCCTTGTGCCCAGTGGAGATA. This probe would detect both known isoforms of TrkA (Dubus et al., 2000). The TrkB probe was directed to nucleotides 402–1393 (GenBank accession number X77251), which codes for much of the extracellular domain of chicken TrkB. The forward primer was TTGTGCTGCCTGGTGCTGGGTT, and the reverse primer was TCCAGCTGAAGGCAGCCATG. This probe is very similar to the probe used by Garner et al. (1996) and should detect all the known TrkB isoforms, including the kinase-truncated isoforms. The TrkC probe was directed to nucleotides 2521–3042 (GenBank accession number NM_205169), which includes the region coding for the intracellular tyrosine kinase domain. The forward primer was ATGCTGCCTATCCGCTGGAT, and the reverse primer was ACAGGTTGTCGTTTTGTGGG. This probe would detect most known TrkC isoforms except for the truncated isoforms of TrkC (Garner et al., 1994).
Antibodies
The primary antibodies we employed in this study (Table 1) fall into one of several categories: 1) antibodies recognizing a variety of transcription factors commonly used to identify different populations of dorsal spinal cord interneurons (Lmx1b, Pax2, Islet1, Lhx1/5, Lhx9, Evx1/2, Engrail1); 2) antibodies used as pan-neuronal markers (TuJ1, HuC/D, and NeuN); or 3) antibodies specific for one of the three Trk receptors and used here to visualize the central projections of specific populations of DRG neurons.
Table 1.
Antibodies used in this study
| Antigen | Immunogen | Source, catalogue or clone number |
Host | Dilution | Cell populations recognized |
|---|---|---|---|---|---|
| Engrail1 | Chick en1, aa66–85, peptide sequence TTNFFIDNILRPDFGC |
DSHB, contributed by T. Jessell and S. Brenner- Morton, clone 4G11 |
mouse | 1:10 | v1 interneurons |
| Evx1/2 | C-terminal peptide from mouse Evx1, aa 407– 416, DQREEVPLTR |
DSHB, contributed by T. Jessell, clone 99.1-3A2 |
mouse | 1:100 | v0 interneurons |
| Islet1 | C-terminal portion of rat Islet1, aa178–349 |
DSHB, contributed by T. Jessell, clone 39.4D5 |
mouse | 1:100 | dI3 interneurons, DRG neurons, motoneurons, sympathetic preganglionic neurons |
| Lhx1/5 | aa1–360 of rat Lim2 | DSHB, contributed by T. Jessell and S. Brenner- Morton, clone 4F2 |
mouse | 1:5 | dI2, dI4, dI6, dILA, v0, and v1 interneurons |
| Lhx9 | peptide from internal region of human Lhx9, aa 185–198 EYPPQLSYTELAAK |
Santa Cruz biotechnology, Cat.# sc-19350 |
goat | 1:100 | dI1 interneurons |
| Lmx1b | Full length mouse Lmx1b |
T. Müller and C. Birchmeier; Max-Delbrück-Centrum for Molecular Medicine. Berlin, Germany |
guinea pig |
1:5,000 | dI5 and dILB interneurons |
| Pax2 | Mouse Pax2 C-terminal domain, aa-188–385 |
Zymed, Cat.# 71–6000 | rabbit | 1:500 | dI4, dI6, v0, and v1 interneurons |
| TuJ1 | neuronal class III beta- tubulin |
Covance, Cat.#MMS-435P | mouse | 1:1,000 | All neurons; cytoskeleton |
| HuC/D | aa240–251 of human HuD |
Molecular Probes, Cat.# A- 21271, clone 16A11 |
mouse | 1:100 | All neurons; cytoplasm |
| NeuN | Purified cell nuclei from mouse brain |
Chemicon, Cat.#MAB377, clone #A60 |
mouse | 1:500 | All neurons; nucleus and cytoplasm |
| TrkA | Extracellular domain of chick TrkA, aa 1–357 |
Dr. F. Lefcort, Montana State University |
rabbit | 1:5,000 | TrkA+ DRG neurons |
| TrkB | Extracellular domain of chick TrkB, aa1–428 |
Dr. F. Lefcort, Montana State University |
rabbit | 1:6,000 | TrkB+ DRG neurons |
| TrkC | Extracellular domain of chick TrkC, aa1–432 |
Dr. F. Lefcort, Montana State University |
rabbit | 1:4,000 | TrkC+ DRG neurons |
The various transcription factor antibodies each specifically recognize the same populations of spinal cord neurons as has previously been described by numerous investigators using other antibodies and in situ hybridization (reviewed by Caspary and Anderson, 2003; Helms and Johnson, 2003; Lewis, 2006). Further, transcription factors tend to be highly conserved across species and, with the exception of the anti-Lhx9 antibody, these antibodies had all previously been shown to detect the homologous chicken protein.
The anti-Lmx1b antibody is directed against the full-length mouse protein, which shares 91% homology with chicken Lmx1b. It specifically labels dI5 and dILB interneurons in the mouse spinal cord, as do other anti-Lmx1b antibodies (Müller et al., 2002 and personal communication), and thus does not cross-react with mouse Lmx1a, which is expressed in the roof plate. In chick spinal cord, it yields the same labeling pattern as described using other anti-Lmx1b antibodies (Muller et al., 2002; Chizhikov and Millen, 2004), including the 50.5A5 monoclonal antibody (DSHB, contributed by T. Jessell), for which the full-length chicken protein was the immunogen. We cannot be certain that the anti-Lmx1b polyclonal antibody we used does not cross-react with chicken Lmx1a, because Lmx1a and Lmx1b are both expressed in roof plate in chick embryos (Chizhikov and Millen, 2004). However, since dI5 and dILB neurons express only Lmx1b, and not Lmx1a, any such cross-reactivity would not confound the results.
The anti-Pax2 antibody is specific for the Pax2 protein and does not cross-react with other Pax family members (Dressler and Douglass, 1992). Immunoblotting, using mouse embryonic kidney lysates, reveals two bands: a major band at 46 kDa (Pax2b) and a minor band at 48 kDa (Pax2a), representing two isoforms of Pax2 from alternative splicing. This antibody also recognizes tumor cells in which Pax2 mRNA could be detected with in situ hybridization (Dressler and Douglass, 1992). Mouse and chicken Pax2 share 93% identical sequences for most of the region (aa204–373) that was used as the immunogen. The pattern of staining we saw using the anti-Pax2 antibody is distinctly different from those revealed by labeling with antibodies against Pax3, 6, and 7.
The anti-Islet1 antibody recognizes both Islet1 and Islet2 (Tsuchida et al., 1994). The sequence of the immunogen (aa178–349 of rat Islet1) is 100% identical to that of chick Islet1, and 80% identical to the corresponding regions of rat and chick Islet2. The antibody labels motoneurons, sympathetic preganglionic neurons, DRG neurons, and dI3 dorsal interneurons during early development, all of which express Islet1, as shown with in situ hybridization (Tsuchida et al., 1994). Since we used this antibody to identify dI3 neurons, the differential expression of Islet1 and Islet2 by some subpopulations of motoneurons, would not confound our analysis.
The anti-Lhx1/5 antibody yields the same labeling pattern as that obtained from in situ hybridization (Tsuchida et al., 1994). The aa1–360 region of rat Lhx5 (NP_620605), which was used as immunogen in generating the antibody, shares 91% identical sequence with chick Lhx5, and 77% with chick Lhx1.
The anti-Lhx9 antibody specifically marks dI1 dorsal interneurons, yielding a pattern of expression identical to that first reported by the Jessell lab using in situ hybridization and an antibody they had generated (Liem et al., 1997; Lee et al., 1998). The immunogen was a 14aa peptide from the internal region of human Lhx9 that shares 100% sequence identity with chick Lhx9 and less than 50% identity with the most closely related LIM-homeodomain protein, Lhx2, making cross-reactivity highly unlikely. However, even if the anti-Lhx9 antibody could detect Lhx2, it would not represent a problem in the experiments described here, since dI1 neurons coexpress Lhx2 and Lhx9 early in development. Importantly, the anti-Lhx9 antibody also does not label interneurons that express other closely related proteins, Lhx1/5 and Lmx1b.
The anti-Evx1/2 antibody recognizes mouse and chick Evx1/2 (Pierani et al., 1999) and specifically detects v0 neurons, consistent with the expression profile obtained from in situ hybridization (Burrill et al., 1997; Matise and Joyner, 1997).
The anti-Engrail1 antibody yields a labeling pattern similar to that of a well-characterized anti-en1 rabbit polyclonal antibody which had been generated by using the conserved 117aa C-terminus of the protein as the immunogen (Davis et al., 1991; Ericson et al., 1997).
We used the TuJ1, HuC/D, and NeuN antibodies to label neurons in different situations. The TuJ1 and HuC/D antibodies label neurons as soon as they begin to differentiate (Lee et al., 1990; Marusich et al., 1994; Burrill et al., 1997). The HuC/D antibody yields intense cytoplasmic labeling and was most useful for visualizing DRG neurons, whereas the NeuN antibody yields nuclear and some cytoplasmic labeling and labeled the small, densely packed neurons in the dorsal spinal cord more distinctly.
The TuJ1 antibody recognizes neuronal class III beta-tubulin, but not the beta-tubulin found in glial cells (Lee et al., 1990). It reveals a 50kDa band in immunoblots prepared using lysates from an immortalized sensory neuroblast cell line derived from mouse otocysts (Lawoko-Kerali et al., 2004).
The anti-HuC/D antibody was developed to study the expression of the Hu family of RNA binding proteins in the developing nervous system of chick embryos (Marusich et al., 1994). This antibody labels NIH3T3 cells transfected with cDNA for chick HuC or HuD, but not HuA, and yields the same pattern of expression in chick embryos as does in situ hybridization. Further, HuC and HuD are expressed only in neurons, starting from the time they begin to differentiate (Wakamatsu and Weston, 1997).
The NeuN antibody was raised against purified cell nuclei from mouse brain and shows reactivity against chick neuronal cells (manufacturer’s datasheet). The antibody recognizes two bands at 46kDa and 48kDa in immunoblots using nuclear extracts from adult mouse brain (Lind et al., 2005). The NeuN antibody stains all neuronal nuclei with exception of mitral cells in the olfactory bulb and Purkinje cells in the cerebellum (Mullen et al., 1992) and is frequently used as a pan-neuronal marker.
The anti-TrkA, anti-TrkB, and anti-TrkC antibodies were each generated against, and are specific for, the extracellular domains of the chicken forms of each of these proteins. Each antibody specifically labels HEK293 cells transfected with cDNA coding for the relevant Trk receptor (Lefcort et al., 1996; Oakley et al., 1997). The anti-TrkC antibody recognizes a 130kDa protein in Western-blots prepared using lysates from COS7 cells transfected with full-length chick TrkC cDNA, but not from mock transfected cells, or COS7 cells transfected with full-length chick TrkA or TrkB cDNA (Lefcort et al., 1996). In addition, the three antibodies mark distinct populations of E10 chick DRG neurons, consistent with the expression profiles obtained by in situ hybridization (Oakley et al., 1997).
Tissue preparation and processing
Tissue preparation and processing was previously described (Honig, 1982; Honig et al., 1998, 2005). All procedures involving animals adhere to NIH guidelines were approved by the Institutional Animal Care and Use Committee at the University of Tennessee Health Science Center. White Leghorn chick embryos were removed from the egg, decapitated, eviscerated, staged according to Hamburger and Hamilton (1951) and fixed in 4% paraformaldehyde, 10% sucrose in 0.1 M phosphate buffer (PB; pH = 7.2–7.4) for 2–6 hours, depending on the size of the embryo. Embryos were subsequently cryostat-sectioned at 20 µm or 30 µm, transverse to the long axis of the spinal cord. The sections were mounted directly, in serial order, onto Superfrost/Plus slides (Fisher), and stored at −80°C until being used.
In situ hybridization
The in situ hybridization protocol was modified slightly from protocols published by Simmons et al. (1989), Schaeren-Wiemers and Gerfin-Moser (1993), and Stern (1998), as described in Honig et al. (2005). Briefly, slides were post-fixed in 4% paraformaldehyde, treated with proteinase K, and acetylated. Hybridization buffer was: 50% formamide, 1× SSC (pH = 7.0), 1× Denhardt’s, 1 mM EDTA, 10% dextran sulfate, 100 µg/ml salmon sperm DNA, 50 µg/ml yeast RNA, 50 µg/ml yeast tRNA, 1% blocking reagent (Roche Applied Science, Indianapolis, IN), and 0.1% SDS. After hybridizing overnight at 65°C, slides were washed first with 50% formamide/ 2× SSC (pH = 7.0), then with 2× SSC, and finally with TBS. Slides were blocked with 0.2% casein in TBS and then incubated overnight with anti-digoxigenin-AP conjugate Fab (Roche Applied Sciences, cat# 11093274910, preabsorbed with chick embryo powder and diluted in TBS with 1% Tween 20 and 0.2% casein) at 4°C and subsequently reacted in nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate. Fluorescent detection used DIG- and/or DNP- labeled riboprobes. Slides were incubated in sheep anti-DIG (Roche Applied Science, cat#1333089) and/or rabbit anti-DNP (Invitrogen, cat#A-6430), and subsequently with appropriate secondary antibodies conjugated to Alexa 488 or 594 (Molecular Probes).
Immunofluorescence procedures
For tissue in which in situ hybridization was combined with immunofluorescence, the immunofluorescent processing was performed after in situ hybridization. For tissue not subjected to previous in situ hybridization, the sections were first permeabilized with 0.5% Triton X-100 in 0.1 M PB for 30 min and blocked with 3% BSA in 0.1 M PB. Sections were incubated in primary antibodies for 2 hours at room temperature or overnight at 4°C. After washing in 0.1 M PB, sections were then incubated in appropriate species-specific secondary antibodies conjugated to Alexa 488, 594, or 647 (Molecular Probes) or with donkey anti-guinea pig Cy3 (Jackson Immunoresearch) for 1–2 hours at room temperature. Slides were coverslipped in Fluoromount-G (SouthernBiotech).
Dorsal root fills
DRG projections into the spinal cord of St. 37 embryos were labeled using a slight modification of methods described previously (Honig and Rutishauser, 1996). In brief, embryos were removed from the egg, placed into oxygenated Tyrode solution, decapitated, eviscerated, and a ventral laminectomy was performed. The ventral roots were cut and the DRGs in the lumbosacral region were exposed. A 15% solution of dextran conjugated to Alexa 488 (10,000 MW, anionic, fixable Molecular Probes #D-22910; dissolved in 0.1 M PB with 0.5% Triton X-100) was pressure-injected using a micropipette with a ~10µm tip. Since uptake of dextran amines is enhanced by damage (Glover et al., 1986), we injected each dorsal root at several points along its width, cut it in the middle of the injected area and let the proximal stump sit in a “ pool” of dye for several minutes before removing the excess. After incubating overnight at room temperature to allow for dye transport, embryos were fixed and processed for immunofluorescence as described above.
Image capture and analysis
Fluorescently labeled tissue sections were viewed with a BioRad MRC 1024 confocal laser-scanning microscope. Bright-field labeling was viewed with an Olympus BH2 microscope and photographed with an Optronics Microfire camera. Confocal images were merged and processed in Adobe Photoshop. Although the sections typically were triple-labeled, in the figures only two channels are shown, for clarity of presentation. Brightness and contrast were modified, and in Fig. 3, extraneous debris and autofluorescent red blood cells in the mesenchyme adjacent to the spinal cord were removed from some of the images.
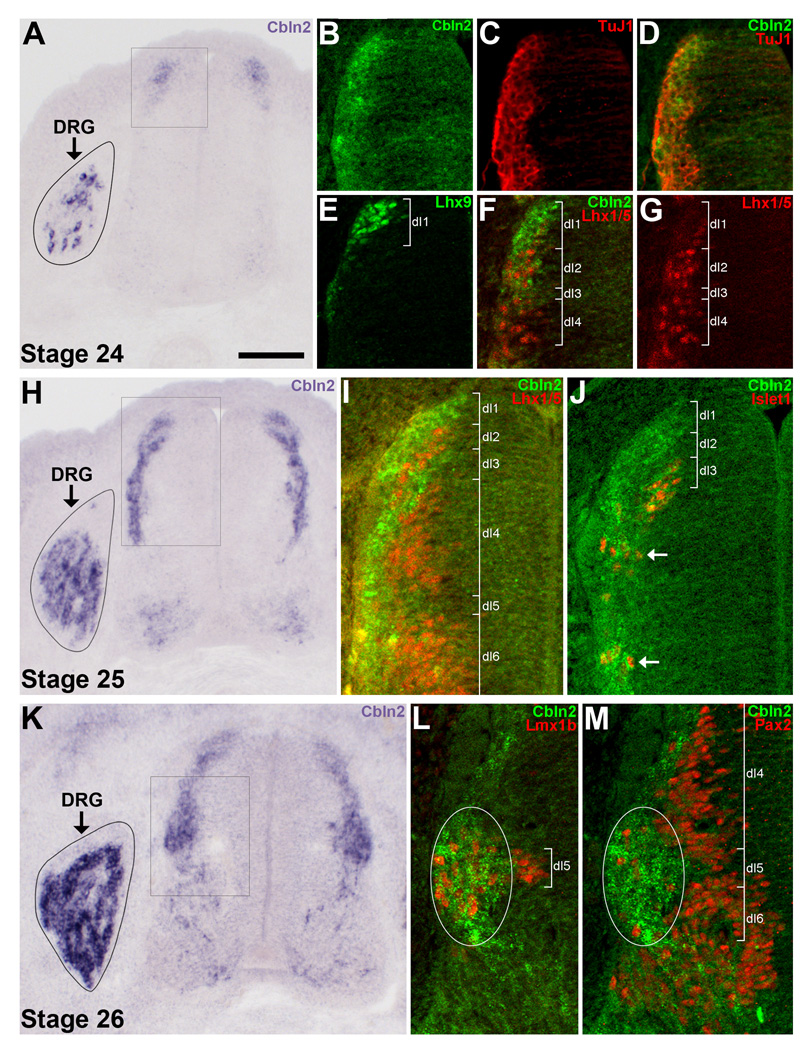
Figure 3. In situ hybridization for Cbln2 in the spinal cord and DRGs at St. 24–26.
A–G) Cbln2 expression at St. 24. A) Cbln2 is expressed by several neurons in the incipient DRGs and at the far dorsal edge of the spinal cord. B–G) Only the dorsal part of the left side of the spinal cord (indicated by the box in panel A) is shown. B–D) Cbln2+ cells at the far dorsal edge of the spinal cord are TuJ1+. E) Lhx9+ dI1 neurons are situated at the far dorsal edge of the spinal cord. F) Cbln2 is predominantly expressed in the region just dorsal to Lhx1/5+ dI2 neurons, by neurons in the same position as the Lhx9+ dI1 neurons shown in panel E. G) The same section as in panel F, showing immunofluorescent staining for Lhx1/5 alone to visualize dI2 and dI4 neurons. H–J) Cbln2 expression at St. 25. H) Cbln2 is expressed by neurons spread along the lateral border of the dorsal half of the spinal cord, by some neurons in the ventral cord, and by an increasing number of DRG neurons. I–J) Only part of the left side of the spinal cord (indicated by the box in panel H) is shown. I) Cbln2 is expressed by a few of the most laterally-situated Lhx1/5+ dI2, dI4 and dI6 neurons. There is a gap in Cbln2 labeling in the dI2 region. J) Cbln2 is expressed by most Islet1+ dI3 neurons, a few of which have already migrated ventrally (arrows). K–M) Cbln2 expression at St. 26. K) Cbln2 is expressed by numerous DRG neurons. In the spinal cord, many dI1 and dI3 neurons (located above the boxed region) express Cbln2, as they do at St. 25, but Cbln2 expression has now expanded to also include a large cluster of neurons (situated within the boxed region). L–M) Only part of the left side of the spinal cord (indicated by the box in panel K) is shown. Cbln2 is expressed by the several Lmx1b+ dI5 neurons and the few Pax2+ dI4/dI6 neurons that are situated within this lateral cluster (white ovals). For all images, dorsal is up. For A, H, and K the DRG on the left side only is shown due to space considerations. For B–D, F–G, I–J, and L–M, in situ hybridization for Cbln2 was followed by processing for immunofluorescence. Note that Cbln2 mRNA is localized to the cytoplasm whereas the transcription factors are expressed in the nucleus, and so double-labeled neurons do not necessarily appear yellow. Magenta-green version available as Supporting Figure 3. Scale bar in A is 100µm for panels A, H, and K and 50µm for panels B–G, I–J, L–M.
Analysis of Cbln2 and Trk expression in the DRGs
To determine which types of sensory neurons express Cbln2, sections were processed for in situ hybridization for Cbln2 and TrkA, TrkB, or TrkC. One probe was visualized with Alexa 488, the other with Alexa 594, and all DRG neurons were revealed by immunofluorescent staining using an anti-HuC/D antibody and an Alexa 647- conjugated secondary antibody. Confocal images through the center of lumbar DRGs were captured with a 20× (NA=0.7) objective, so that the entire cross-section of the DRG could be visualized. ImageJ was used to separately threshold the images for Cbln2 and for Trk labeling, and show them individually over the HuC/D version of the image to reveal pixels above the threshold. Neurons, as revealed by HuC/D labeling of the cytoplasm, with >4 pixels above threshold for the in situ probe were counted by eye using the Cell Counter ImageJ plugin. The markers from the Cbln2/HuC/D image and those from the Trk/HuC/D image were then compared to determine which neurons expressed one marker or the other, and which neurons expressed both. Visual inspection of additional images captured with a 40× oil immersion objective (NA = 1.35) confirmed the extent of Cbln2/Trk coexpression.
In examining sections processed for in situ hybridization, we observed considerable variation in the absolute numbers of Cbln2+, TrkB+, and TrkC+ neurons. In the sections we analyzed in detail, we found corresponding variation in the percentages of Cbln2+ neurons expressing TrkB (32–47%) and of Cbln2+ neurons expressing TrkC (43–53%). This variation is likely to be the result of differences between DRGs at different segmental levels and possibly also differences within individual DRGs. Regardless, the complement of Cbln2+ sensory neurons would seem to be roughly evenly distributed between those expressing TrkB and those expressing TrkC.
Analysis of Cbln2 expression in dorsal horn interneurons
To determine which types of dorsal horn interneurons express Cbln2, sections were processed for in situ hybridization for Cbln2 followed by double immunofluorescent labeling for Lmx1b and Pax2. Confocal images were captured with a 20× (NA=0.7) objective and subsequently overlaid with an outline indicating the boundaries of laminae I – IV, as could best be determined using the criteria described in the Results. Six sections from four different embryos were analyzed in detail. The number of Lmx1b+ and Pax2+ nuclear profiles (and hence, neurons) were counted using ImageJ and the Image-based Tool for Counting Nuclei (ITCN) plugin obtained from the ImageJ website. To quantify the extent of Cbln2 expression, although the sections themselves were triple-labeled, only two channels were combined at a time, with Cbln2 in green and the neuronal marker in red. Neurons expressing Cbln2 were identified using the Colocalization Highlighter ImageJ plugin and separately thresholding each of the channels. The macro detects sites of spatial overlap between two channels and displays them as white pixels. Because the transcription factors Lmx1b and Pax2 are localized to the nucleus, whereas Cbln2 mRNA is in the cytoplasm, white pixels appear only in areas where the nuclear labeling and the cytoplasmic labeling are very closely apposed in the x- and/or y-axes or overlie one another in the z-axis. Given the ~6µm depth of the optical section relative to the size of these dorsal horn interneurons (nuclear diameters 5–7µm; cell diameters ~7–10µm), any overlying Cbln2 labeling is far more likely to be localized to the cytoplasm of that particular neuron rather than to a neuron whose nucleus lies out of the image plane. Most neurons that appeared by eye to be Cbln2+ were characterized by 10–50 white pixels within their 50–100 pixel area. The presence of 1 or 2 white pixels per neuron seemed to be spurious and related to the slight imprecision inherent to thresholding. By comparison to the original, unaltered images, we decided to use 4 white pixels as the cut-off for considering a neuron to be Cbln2+. Examples of this analysis are shown in Supporting Fig. 2. We realize that the actual number of coexpressing neurons detected with this approach will depend on the number of white pixels used as the cut-off value and the precise level used for thresholding the images of Cbln2 labeling. However, since our goal was to determine the relative proportions of Cbln2+ neurons in the different laminae and the relative proportions of Lmx1b+ neurons and Pax2+ neurons that express Cbln2, rather than the absolute numbers of neurons, the use of a slightly different cut-off or threshold level would have not altered the overall results. Cbln2+ neurons were marked and counted. Comparisons between laminae were performed using a nonparametric, two-tailed Mann-Whitney U test (http://elegans.swmed.edu/~leon/stats/utest.cgi).
RESULTS
Characterization of the chicken CBLN2 transcript
We assembled the BB180 clone and related ESTs into a contig (Fig.1; Genbank accession number GU189513, see Material and Methods). The assembled contig spans a 6kb long genomic region on chromosome 2, from 94978728 to 94984780, according to the most recent version (May, 2006) of the chicken genome assembly (BLAT program, http://genome.ucsc.edu/cgi-bin/hgBlat). This contig is separated into five exons, with the predicted introns starting with GT and terminating with AG, consistent with the canonical intron structure. There is, however, an un-sequenced gap in the region between exons 2 and 3. RT-PCR was performed to amplify a 1.8kb fragment of this contig covering the first coding exon (putative exon 2) and the original 180bp clone (putative exon 5). This PCR fragment was sequenced and found to be identical in sequence to the comparable part of our assembled contig. We have not been able to amplify the first 380bp sequence in the assembled contig (the 5” end of exon 1), probably due to its high GC content.
Analysis of the assembled contig reveals a 636nt-long open reading frame encoding a 212aa protein. This protein is highly homologous to Cbln2 from a wide range of vertebrate animals, including human, mouse, frog, and fish (Fig 2A). Chicken Cbln2 also contains the two asparagine residues for N-glycosylation, and the two cysteine residues for putative disulfide bond formation that are conserved in the Cbln family in mammals (Bao et al., 2005; Iijima et al., 2007). Its C-terminus is predicted to contain a 140aa-long C1q domain, using the SMART program (http://smart.embl-heidelberg.de/). The chicken Cbln2 C1q domain is 100% identical to that of the Cbln2 homologues in human, monkey, cow, mouse, and rat, and shares 82% and 76% identity with those of chicken Cbln1 and Cbln4, respectively (Fig. 2B). Thus, our assembled contig appears to represent the chicken homologue of Cbln2.
Early expression of Cbln2
To characterize the expression of Cbln2 during early spinal cord and DRG development, we examined embryos starting at St. 24 (4 days of development). At this time, motoneuron axons are beginning to exit the limb plexuses and extend along specific peripheral nerve pathways (Lance-Jones and Landmesser, 1981; Tosney and Landmesser, 1985). All motoneurons have been generated (Hollyday and Hamburger, 1977) and commissural axons have crossed the floor plate (Stoeckli and Landmesser, 1995), but otherwise the spinal cord is very immature. Similarly, the axons of the oldest DRG neurons have reached the plexus in the periphery (Landmesser and Honig, 1986) and the dorsal funiculus centrally (Davis et al., 1989), but sensory neurons are still being generated (Carr and Simpson, 1978).
At St. 24, Cbln2 is expressed by a small number of cells in the incipient DRGs that have a neuronal morphology (Fig. 3A) and express the neuronal markers HuC/D and TuJ1 (data not shown). The number of Cbln2+ neurons then rapidly increases (Fig. 3H), concomitant with the generation and differentiation of additional neurons in the DRGs (Carr and Simpson, 1978; George et al., 2010). By St. 26, Cbln2 is expressed by many, but not all, neurons in the core of the DRG, and few, if any, at the dorsomedial pole where newly generated neurons predominate (Fig. 3K).
Cbln2 is also expressed in the spinal cord from St. 24 on. As shown in Figs. 3H and K, Cbln2+ neurons are sometimes present ventrally, in the position of motoneurons and sympathetic preganglionic neurons, which at these times are indistinguishable (Prasad and Hollyday, 1991). Based on the spatial pattern of Cbln2 expression a few days later (see the next section of the Results), when motoneurons are clustered in the ventral horn and separate from sympathetic preganglionic neurons in the column of Terni, it is likely that the ventrally located Cbln2+ neurons at St. 24–26 include both sympathetic preganglionic neurons and motoneurons.
In the dorsal spinal cord, the pattern of Cbln2 expression changes rapidly between St. 24 and St. 26. At St. 24, a small number of TuJ1+ and HuC/D+ neurons located at the far dorsal edge of the spinal cord express low levels of Cbln2 (Fig. 3A–D; HuC/D expression not shown). By St. 25, the intensity and extent of staining for Cbln2 increases, with Cbln2 being expressed by neurons that are spread along the lateral border of the dorsal half of the spinal cord (Fig. 3H). By St. 26, the number of Cbln2+ neurons further increases, and includes a large cluster of cells in an intermediate position (Fig. 3K).
During spinal cord development, neurons are generated within the ventricular zone and then migrate to the lateral edge of the cord. In St. 24 chick embryos, as in E10 mice, six dorsal interneuron populations (dI1–6) and five ventral neuron populations (v0–v3 and motoneurons) are present, each occupying a discrete dorsal-ventral position and expressing a specific complement of transcription factors (reviewed by Caspary and Anderson, 2003; Helms and Johnson, 2003; Lewis, 2006). To determine the identity of the Cbln2+ neurons in the spinal cord with respect to these populations, we processed some sections first for in situ hybridization (using fluorescence detection) and subsequently for immunofluorescence using antibodies against transcription factors specific for these neuronal populations.
The first dorsal interneurons to express Cbln2 are Lhx9+ dI1 neurons (Fig. 3E–F). Slightly later, Cbln2 is also expressed by most Islet1+ dI3 (Fig. 3J) and Lmx1b+ dI5 neurons (Fig. 3L). Only a few of the more lateral Lhx1/5+ dI2, dI4, and dI6 neurons express Cbln2 (Fig. 3F–G, I). Cbln2 expression appears not to extend into the ventral part of the spinal cord at St. 25 (Fig. 3H). Since Lhx1/5 also labels v0 and v1 interneurons, to examine this directly, we stained some sections for the v0 interneuron marker, Evx1, and other sections for the v1 interneuron marker, Engrail1. We found that Cbln2 is not expressed by either v0 or v1 interneurons (data not shown), and thus cannot be expressed by v2 and v3 interneurons, which are situated even more ventrally in a territory that is also unlabeled. At St. 26, Cbln2 expression is maintained in the same types of dorsal interneurons as at St. 25, primarily dI1, dI3, and dI5 neurons, and has expanded to include a large cluster of neurons (Fig. 3K). This cell cluster consists of numerous Lmx1b+ dI5 and a few Pax2+ dI4 and dI6 neurons (Fig. 3L–M; Supporting Fig. 1). Cbln2 is not expressed by the more medial Lmx1b+ and Pax2+ neurons, but rather only by those neurons that have already migrated laterally. In addition, while some Cbln2+ neurons are now located in the ventral part of spinal cord, these appear to primarily be dI4 and/or dI6 interneurons (that have begun to migrate ventrally; see Discussion), sympathetic preganglionic neurons, and motoneurons, rather than v0 –v3 neurons. DILA and dILB neurons first appear at late St. 26, are few in number until St. 28, and are still being generated after St. 30; accordingly, their expression of Cbln2 is described in the following section.
Expression of Cbln2 at St. 30
We next characterized the expression of Cbln2 in the spinal cord and DRGs at St. 30–31 (6½–7days), after motoneurons and sensory neurons establish appropriate projections in the limb. As shown in Fig. 4A and C, many Cbln2+ neurons are present in the spinal cord, with the majority being clustered in two regions: (1) the dorsal horn, which receives extensive input from DRG neurons, and (2) the column of Terni, which contains sympathetic preganglionic neurons in birds and extends the length of the thoracic region and into the most rostral one or two lumbar segments. Interestingly, neurons in the sympathetic chain ganglia also express Cbln2 (Fig. 4B). In addition, Cbln2+ motoneurons are found in some sections, sometimes scattered, other times clustered, and typically in the medial motor column or in the lateral part of the lateral motor column (Fig. 4C). Many of the Cbln2+ motoneurons are located in the part of the lateral motor column corresponding to the iliotibialis motoneuron pool (Landmesser, 1978; Lin et al., 1998), but it seems unlikely that all iliotibialis motoneurons are Cbln2+. Further, some Cbln2+ motoneurons are clearly located outside of the region occupied by iliotibialis motoneurons.
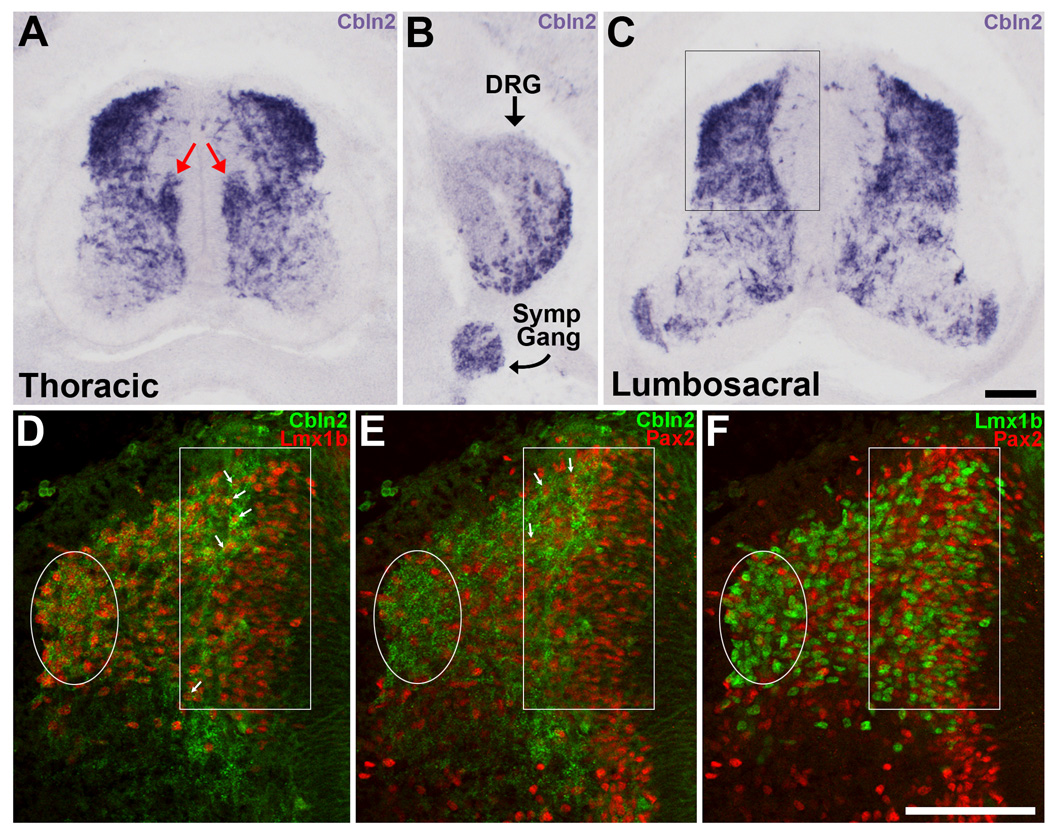
Figure 4. In situ hybridization for Cbln2 in the spinal cord at St. 30.
A) At thoracic levels, Cbln2 is heavily expressed in the dorsal part of the spinal cord and by sympathetic preganglionic neurons in the column of Terni (red arrows). B) In the same section as in (A), Cbln2 is expressed in the DRGs and in the sympathetic chain ganglia. Medial is to the left. C) In lumbosacral segments, Cbln2 is heavily expressed in the dorsal half of the spinal cord and by some motoneurons. D–F) The dorsal part of the left side of the spinal cord (indicated by the box in panel C) is shown. The same section is shown in all three panels. The lateral cluster of dI4–6 neurons has migrated dorsally (enclosed in white oval) and is now situated at the dorsolateral edge of spinal cord. Numerous dILA and dILB neurons are now present and located medially (white rectangle). D, E) Some of the more lateral Lmx1b+ dILB and Pax2+ dILA neurons express Cbln2 (arrows). F) Lmx1b+ dILB and Pax2+ dILA are intermixed in a salt-and-pepper-like manner. Magenta-green version available as Supporting Figure 4. Scale bar, 100µm in C is for panels A, B, and C; in F is for panels D, E and F.
The pattern of Cbln2 expression in the dorsal part of the spinal cord at St. 30 is very different than that observed at St. 26. Cbln2 is intensely expressed by the lateral cluster of Lmx1b+ and Pax2+ dI4–6 neurons that was first noted at St. 26, and has since migrated dorsally along the lateral edge of the spinal cord (Fig. 4D–F). Cbln2 is also expressed more medially in the dorsal spinal cord. A second phase of neurogenesis starts at about St. 27 (E11.5 in mouse) and continues for several days. It gives rise to two additional populations of dorsal interneurons called dILA and dILB, which are intermixed and express Pax2 and Lmx1b, respectively (Gross et al., 2002; Muller et al., 2002, 2005; Wildner et al., 2006). By St. 30, considerable numbers of dILA and dILB neurons have been generated, the oldest of which have begun to migrate laterally away from the ventricular zone. Some of the older, more lateral dILA and dILB neurons express Cbln2 (Fig. 4D, E), whereas the younger, more medial dILA and dILB neurons do not.
A substantial number of sensory neurons also express Cbln2, particularly those in the ventral and lateral parts of the DRGs (Fig. 5A). This pattern of labeling suggests that Cbln2 expression may be restricted to a particular subpopulation of sensory neurons. In the chick, two classes of DRG neurons can be distinguished from about St. 29 until about St. 36: older, larger neurons in the ventrolateral (VL) region and younger, smaller neurons in the dorsomedial (DM) region (Hamburger and Levi-Montalcini, 1949). Further, although nearly all DRG neurons initially express more than one type of Trk receptor (Rifkin et al., 2000), by St. 30 most neurons express a single Trk receptor and neurons expressing different Trk receptors tend to be spatially segregated within the DRGs (Fig. 5B–D; see also Oakley et al., 1997; Rifkin et al., 2000). TrkA is expressed by small neurons in the DM part of the DRGs, TrkC by large neurons situated at the lateral edge of the DRGs, and TrkB by large neurons that are intermixed with or medial to the TrkC+ neurons.
Figure 5. Cbln2 is expressed by TrkB+ and TrkC+ DRG neurons, but not by TrkA+ DRG neurons. In situ hybridization on sections from St. 30 embryos.
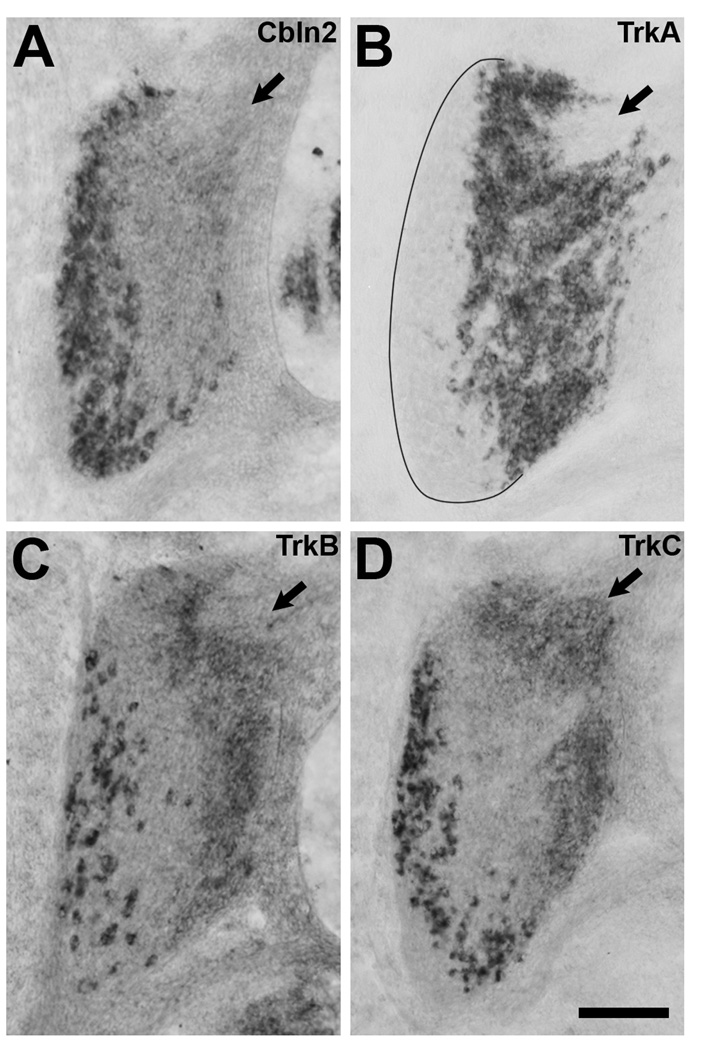
A) Cbln2 is expressed primarily by large neurons in the ventral and lateral parts of the DRG. B) TrkA is expressed by small neurons in the dorsomedial part of the DRG. C) TrkB is expressed by large neurons situated amongst and slightly medial to the TrkC+ neurons. D) TrkC is expressed by large neurons situated at the lateral edge of the DRG. Low levels of TrkB and TrkC expression in small neurons in the dorsomedial region were not considered in our analysis. Some of the small neurons in the dorsomedial region expressing low levels of TrkB or TrkC express high levels of TrkA and were therefore considered to be TrkA+ in our analysis. The dorsomedial pole of each DRG is marked with an arrow. Dorsal is toward the top, lateral to the left. Scale bar, 100µm.
To determine the Trk receptor expression profile of Cbln2+ DRG neurons, we carried out in situ hybridization on sections through the lumbosacral region using two different fluorophores to visualize two kinds of mRNA in the same section. (The small neurons at the DM pole that express high levels of TrkA together with low levels of TrkB and/or TrkC were not considered in our analysis; see also Lefcort et al., 1996). We found that nearly all TrkB+ neurons (95.9% n=2 sections) and nearly all TrkC+ neurons (96.25% n=2 sections) express Cbln2. In contrast, extremely few Cbln2+ cells express TrkA (3.4% n=3 sections). Examination of three additional sections for each Trk/Cbln2 combination, showing only one TrkB or TrkC neuron per section does not express Cbln2 and only one TrkA neuron per section does, confirmed these results. In addition, in sections labeled for TrkA and Cbln2, the only neurons that do not express either TrkA or Cbln2 are located at the DM pole, where progenitor cells are found and neurons are still being generated (George et al., 2010). Thus, Cbln2 is expressed by both TrkB+ and TrkC+ sensory neurons, but not by TrkA+ neurons.
Expression of Cbln2 in the dorsal horn at St. 37
Our observation that Cbln2 is co-expressed by sympathetic preganglionic neurons and their synaptic targets in the sympathetic chain ganglia, led us to consider the possibility that the neurons expressing Cbln2 in the dorsal spinal cord correspond to the central targets of TrkB+ and TrkC+ DRG neurons. To do this, we examined Cbln2 expression in lumbar segments in embryos at St. 37 (11 days), when the spinal cord is more mature. As shown in Fig. 6A, Cbln2 is most robustly expressed in the dorsal horn and numerous large Cbln2+ neurons are present in the intermediate part of the spinal cord.
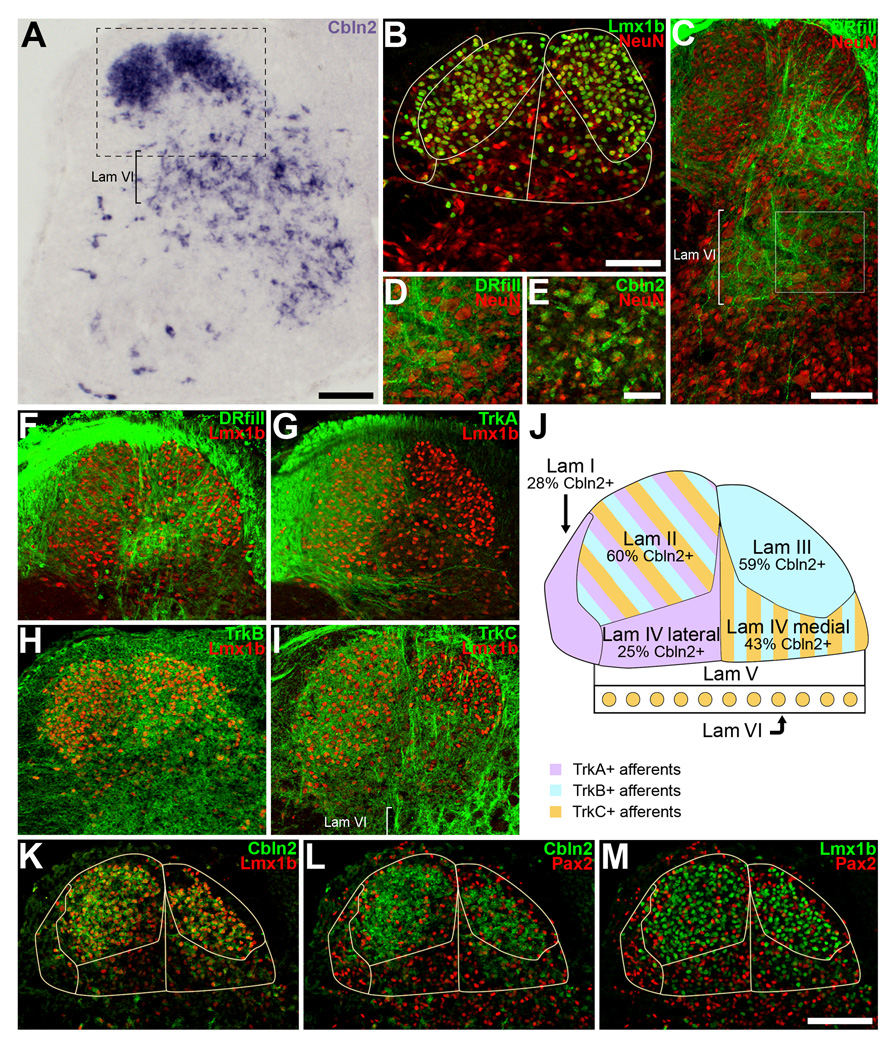
Figure 6. At St. 37, Cbln2 is expressed in regions of the dorsal and intermediate spinal cord where TrkB+ and TrkC+ axons arborize.
A) Cbln2 is most heavily expressed in the dorsal horn and in lamina VI. B) Laminae I–IV can be distinguished by immunofluorescent staining for Lmx1b and NeuN. See text for description and panel J for lamina identification. The area shown is equivalent to that indicated by the box in panel A. C) Sensory axons arborize most extensively in lamina IV and lamina VI. The dorsal roots (DR) were filled with Alexa 488-dextran. D) Higher magnification view of the box outlined in panel C. Sensory axons arborize amidst the large neurons in lamina VI. E) Cbln2 is expressed by large neurons in lamina VI. The area shown is from a different embryo but is similar to that shown in panel D. F–I) Sensory axon projections into the dorsal spinal cord. Sections were also labeled for Lmx1b to aid in distinguishing laminae. F) Sensory axons filled with Alexa 488-dextran arborize in laminae I, II, III, and IV. G) TrkA+ afferents terminate in laminae I and II, and the lateral part of lamina IV. The same section as in panel F is shown. H) TrkB+ afferents arborize primarily in lamina III and the medial part of lamina IV, and to a lesser extent, in the dorsolateral part of lamina II. I) TrkC+ afferents arborize in lamina II, the medial part of lamina IV, and in lamina VI. TrkC+ axons cross lamina III on their way to lamina IV, but it is not clear if any of them terminate in lamina III. In some embryos, light labeling for TrkC and for TrkB could be seen in lamina I. J) Schematic overview of laminae I–VI. Colors indicate where TrkA+, TrkB+, and TrkC+ axons arborize. The percentage of neurons in each lamina that expresses Cbln2 is indicated. K–L) Cbln2 is most abundant in lamina II and lamina III and expressed at lower levels in lamina I and lamina IV. K) Many Lmx1+ neurons express Cbln2. L) Some, albeit fewer, Pax2+ neurons express Cbln2. M) Lmx1+ and Pax2+ neurons are intermixed in laminae I–IV. Magenta-green version available as Supporting Figure 5. Scale bars, 100µm in A, C, M; 50µm in B and E. Scalebar in M also applies to panels F–I, K–L. Scale bar in E also applies to panel D.
To characterize the pattern of Cbln2 expression in more detail, we then sought to distinguish specific laminae, and identify the central targets of TrkB and TrkC afferents with respect to these laminae. This entailed the use of several approaches. First, we used the NeuN antibody to label all neurons to help us apply the criteria that Martin (1979) had established for adult chick lumbar spinal cord based on Rexed’s scheme for cat (Fig. 6B). The arrangement of laminae I–IV in the chick differs from the more typical arrangement seen in mammals, in which these laminae are stacked dorsoventrally. In the chick, laminae II and III are oval-shaped and lie side by side, with lamina II being lateral and lamina III being medial, separated by sensory axons coursing ventrally from the dorsal funiculus (Brinkman and Martin, 1973; Martin, 1979; see also Woodbury and Scott, 1991; Eide and Glover, 1997). Lamina II and lamina III each contain many small, densely-packed neurons and can be most readily identified by the numerous Lmx1b+ neurons present. Lamina I is a thin layer of cells that surrounds lamina II laterally and ventrally, and often includes some displaced neurons that are situated in the white matter. Lamina IV is triangular in shape, situated between and ventral to laminae II and III, with small and medium-sized neurons that are less densely packed than in laminae II and III, but more densely packed than in the cell-poor lamina V. Lamina VI is a cell-rich band lying ventral to lamina V, and characterized by the presence of very large neurons.
We next wanted to identify the central targets of TrkB+ and TrkC+ DRG neurons. Although TrkB+ and TrkC+ axons in the spinal cord have previously been visualized by immunolabeling (see Discussion), detailed descriptions of their projections relative to laminar boundaries in the chick appear to be lacking. To elucidate the pattern of central projections, we examined sections of triple-labeled tissue that had been prepared using various combinations of fluorescent dextran labeling of the dorsal roots, immunofluorescent labeling for TrkA, TrkB, or TrkC, and immunofluorescent labeling for Lmx1b or NeuN. As shown in Fig. 6C, D, and F, sensory axons (labeled by dorsal root fills) arborize in laminae I, II, III, IV, and VI, and some course to the lateral motor column. We found that TrkC+ axons arborize primarily in lamina II, the medial part of lamina IV, lamina VI and the lateral motor column (Fig. 6I). TrkB axons arborize in dorsolateral lamina II, lamina III and the medial part of lamina IV (Fig. 6H). For TrkB and TrkC, we sometimes observed light labeling in lamina I, which is probably due to low levels of TrkB and TrkC expression in some of the TrkA+ neurons. We further found that TrkA axons arborize in laminae I and II, as expected, but also that they extend into the lateral part of lamina IV (Fig. 6G), consistent with the results from DiI labeling of cutaneous nerves (Eide and Glover, 1997). Thus, our results show that lamina II receives projections from TrkB+ and TrkC+ axons (not just TrkA+ axons), lamina III receives projections almost exclusively from TrkB+ axons, and medial lamina IV receives projections from both TrkB+ and TrkC+ axons. In contrast, lamina I and lateral lamina IV receive their input mainly from TrkA+ neurons (Fig. 6G).
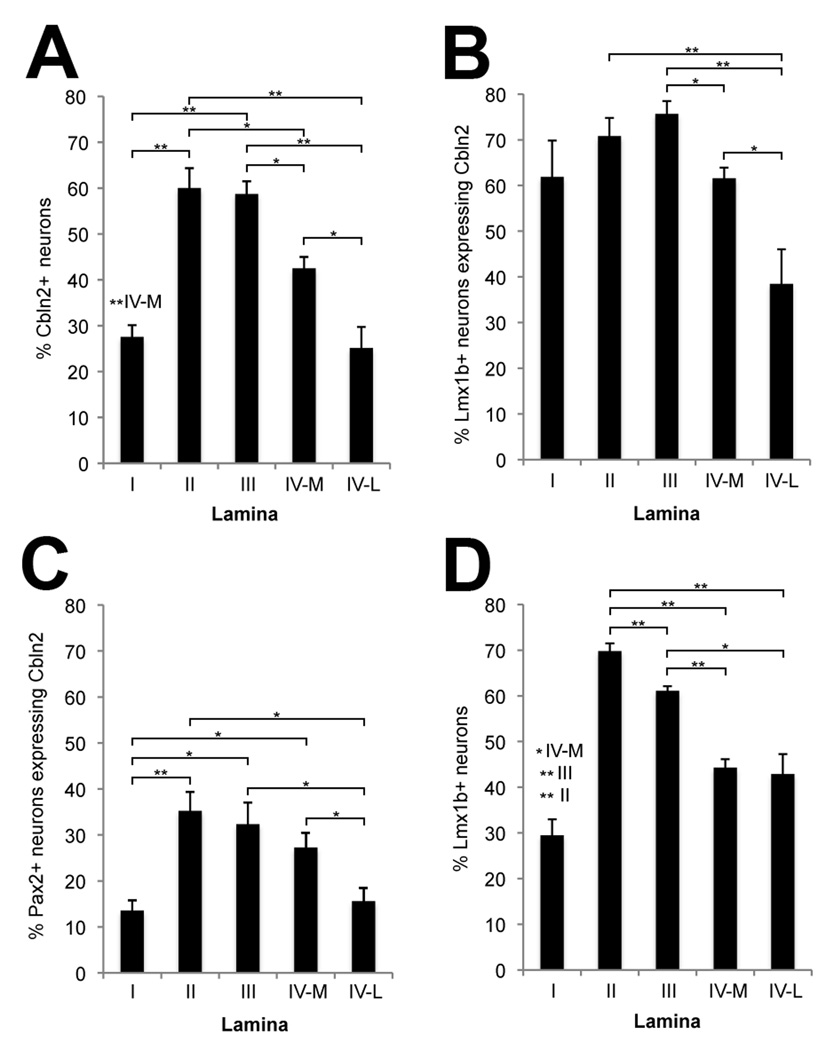
We then processed sections for fluorescence detection of Cbln2 mRNA followed by immunofluorescence for NeuN, Lmx1b and/or Pax2. As shown in Fig. 6E, Cbln2 is expressed by large neurons in lamina VI that receive extensive input from TrkC+ sensory axons. Dorsally, Cbln2 expression appears to be the greatest in lamina II and lamina III, and more moderate in lamina IV. Because the density of neurons varies among laminae, to quantify the extent of Cbln2 expression, we determined the percentage of Cbln2+ neurons in each lamina. Given that medial lamina IV receives TrkB and TrkC input whereas lateral lamina IV receives TrkA input, we considered the two parts of lamina IV separately for this analysis. We found that Cbln2 is expressed by more neurons in lamina II (60%) and lamina III (59%) than in medial lamina IV (42%), and by even fewer neurons in lamina I (28%) and in lateral lamina IV (25%) (Figs. 6J–L, 7A). Thus, proportionally more neurons in regions that receive substantial TrkB and TrkC input (laminae II, III, medial IV) express Cbln2 than in regions that do not receive TrkB and TrkC input (lamina I and lateral IV).
Figure 7. Cbln2 is mainly expressed in lamina II and lamina III of the dorsal horn and more frequently by the excitatory Lmx1b+ neurons than by the Pax2+ inhibitory neurons in dorsal horn.
Cbln2+ neurons in the dorsal horn were counted in six sections that were processed first for in situ hybridization for Cbln2, and then for immunofluorescence to visualize Lmx1b+ and Pax2+ neurons. A) The percentage of neurons that are Cbln2+ is greatest in lamina II and lamina III, less in lamina IV medial, and lowest in lamina I and lamina IV lateral. B) The percentage of Lmx1b+ neurons that are Cbln2+ is lowest in lamina IV lateral. C) The percentage of Pax2+ neurons that are Cbln2+ is lowest in lamina I and in lamina IV lateral. D) The percentage of neurons that are Lmx1b+ is lowest in lamina I and is lower in lamina IV medial and lamina IV lateral than in lamina II or lamina III. Values shown are mean ± standard error of the mean. Statistically significant differences between laminae were determined using two-tailed Mann-Whitney U tests and are indicated either by brackets indicating each comparison or, to minimize excessive crowding, above the bars showing the lamina I values. Probability is indicated by the number of asterisks (*P ≤ 0.05; ***P ≤ 0.005). The significance levels (p values) and the statistic (U) values for these comparison are provided in Supporting Table 1.
Laminae I–IV are each composed of a mixture of GABAergic inhibitory interneurons and glutamatergic excitatory interneurons, which can be distinguished by their (continued) expression of Pax2 and Lmx1b, respectively (Cheng et al., 2004, 2005). To determine whether Cbln2 is expressed by one or both types of interneurons, we combined in situ hybridization for Cbln2 with double immunofluorescent staining for Pax2 and Lmx1b. As shown by the images in Fig. 6K–L and quantified in Fig. 7 (also see Supporting Table 1), Cbln2 is more frequently expressed by Lmx1b+ neurons than by Pax2+ neurons. The overall percentage of Lmx1b+ neurons expressing Cbln2 for laminae I–IV is two times greater than that for Pax2+ neurons (67.8% vs 26.6%; P ≤ 0.0001), and a higher proportion of Lmx1b+ neurons than of Pax2+ neurons express Cbln2 in each lamina (compare panels B and C in Fig. 7). Thus, Cbln2 is expressed preferentially by excitatory interneurons in the dorsal horn.
We also noticed that the different laminae varied in the relative numbers of excitatory vs inhibitory interneurons they contain. Laminae II and III are composed of a higher percentage of Lmx1b+ neurons (69.8% and 61.1%) than is lamina IV (44.3% for medial lamina IV, 42.9% for lateral lamina IV), and lamina I has the lowest percentage (29.5%) (Fig. 7D). This difference may contribute to, but cannot be solely responsible for, the overall laminar differences we found in the extent of Cbln2 expression. For example, the medial and lateral parts of lamina IV are similar in their relative proportions of excitatory and inhibitory interneurons, but the overall percentage of Cbln2+ neurons in lamina IV medial is significantly greater than in lamina IV lateral (42.5% vs 25.1%, P ≤ 0.025; Fig. 7A). In addition, for lamina I, the percentage of inhibitory interneurons expressing Cbln2 is significantly smaller than that percentage for laminae II (13.5% vs 35.2%, P ≤ 0.004) and III (13.5% vs 32.3%, P ≤ 0.011; Fig. 7C).
DISCUSSION
The Cbln family of C1q domain proteins is highly conserved among mammals (Tang et al., 2005; Yuzaki, 2008). Tang et al. (2005) have previously noted that 96–100% of the amino acid sequences for Cbln1, Cbln2, and Cbln4 are identical between mice and humans. We find that Cbln1, Cbln2, and Cbln4 are present in chicken and are 88–90% identical to their human homologues. Further, Cbln1, Cbln2, and Cbln4 are present in zebrafish (Danio rerio, Cbln1 Genbank accession number NP_001002520, Cbln2 NP_001092208, Cbln4 NP_001103861), and frog (Xenopus laevis, Cbln1 NP_001088672, Cbln2 NP_001080811, and Cbln4 NP_001079919). In contrast, Cbln3 appears to be absent in fish, frog, and chicken, and although it has been identified in all known mammalian genomes, it shows less conservation between mouse and human (88%; Tang et al., 2005) than do the other Cblns. Thus, as has previously been noted (Yuzaki, 2008), Cbln3 is indeed likely to be the last member of the Cbln family to have emerged during evolution. Interestingly, Cbln3 differs from Cbln1, Cbln2, and Cbln4 in a few additional respects. Cbln3 expression is restricted to only two areas in the mouse brain (cerebellum and dorsal cochlear nucleus) and first appears at postnatal day 10, whereas the other Cblns are expressed more widely throughout the brain and can first be detected much earlier, at E10 (Pang et al., 2000; Miura et al., 2006). Finally, Cbln3 can only be secreted from cells when it is co-expressed with Cbln1 (Bao et al., 2006), whereas Cbln1, Cbln2, and Cbln4 can each be secreted as homomers (Pang et al., 2000; Bao et al., 2005; 2006; Iijima et al., 2007; J. Morgan, personal communication).
Here we show that Cbln2 is expressed by specific subpopulations of neurons in the DRGs and in the spinal cord. For both structures, Cbln2 expression starts shortly after neurons are generated and continues past the time they establish their connections. In this respect, Cbln2 expression mirrors that of Cbln1, which is present in both granule cell progenitors and mature granule neurons (Wei et al., 2007). During the course of development, the spatial distribution of Cbln2+ neurons in the spinal cord changes considerably, as Cbln2+ neurons migrate within the spinal cord. Further, we show that Cbln2 is frequently expressed by populations of synaptically connected neurons and go on to discuss the possible implications of this finding.
Cbln2 expression in the spinal cord
From St. 24 on, Cbln2 is expressed by some neurons located in the ventral part of the cord. It is likely that these correspond to both motoneurons and sympathetic preganglionic neurons, which are not readily distinguishable at early stages. Motoneurons and sympathetic preganglionic neurons are intermingled as they are generated and migrate laterally away from the ventricular zone, and both types of neuron express Islet1 (Thaler et al., 2004) and choline acetyltransferase. Sympathetic preganglionic neurons undergo a secondary migration starting at about St. 27, toward their final destination in the column of Terni, situated immediately adjacent to the central canal (Prasad and Hollyday, 1991). Indeed, at St. 30, Cbln2 is strongly expressed in the column of Terni. In addition, some motoneurons express Cbln2 though we could not detect a pattern as to which were Cbln2+ and which Cbln2−. Thus, given that sympathetic preganglionic neurons are situated only in thoracic and upper lumbar segments, most of the ventrally located Cbln2+ neurons in those segments at St. 24 are probably sympathetic preganglionic neurons, whereas those in LS2 and more posteriorly are likely to be motoneurons.
In the dorsal part of the spinal cord, Cbln2 is expressed by the most dorsal of the six classes of early developing dorsal interneurons, the dI1 interneurons, by St. 24. This pattern of expression is consistent with a recent study characterizing cells derived from dP1 Atoh+ progenitors (from which dI1 interneurons arise) showing that their spatial distribution is similar to the pattern of Cbln2 expression in the mouse spinal cord at E12.5 (Miesegaes et al., 2009). Later in development, dI1 neurons migrate ventrally to ultimately settle in the deep dorsal horn and project to the cerebellum (Bermingham et al., 2001). Accordingly, at St. 37 we found a number of large neurons expressing Cbln2 in lamina VI. We assume that at least some of these Cbln2+ neurons are dI1 interneurons, although we were not able to successfully label them for the dI1 marker, Lhx9, in older embryos.
After St. 24, Cbln2 expression gradually extends ventrally and ultimately includes the later generated dILA and dILB populations. At St. 26–30, Cbln2 is expressed predominantly by dI3, dI5, and dILB neurons, to some extent by dI2, dI4, dI6, and dILA neurons, but not by v0, v1, v2, or v3 neurons. Several days later, when the spinal cord has matured, the vast majority of Cbln2+ cells are clustered together in lamina II and lamina III, while additional Cbln2+ cells are found scattered primarily in lamina IV and lamina VI (in addition to expression in the column of Terni and in the lateral motor column, as discussed above).
The dramatic changes in the spatial pattern of Cbln2 expression between St. 26 and St. 37 seem to reflect the migration that the various types of neurons undergo within the spinal cord, rather than changes in Cbln2 expression per se. DI3 neurons are known to migrate to the deep dorsal horn (Gross et al., 2002). Indeed, at St. 37, we found Islet1+ dI3 interneurons in lamina VI, some of which express Cbln2. Both dI5 neurons and dILB neurons are identified by their expression of Lmx1b. DILB neurons migrate to the dorsal horn, whereas dI5 neurons are thought to migrate into the ventral horn (Gross et al., 2002; Muller et al., 2002; Qian et al., 2002; Ding et al., 2004). However, we did not detect any Lmx1b+ neurons in the ventral half of the spinal cord in embryos examined at St. 30–37. Rather, Lmx1b+ neurons are located primarily in laminae II–IV at St. 37, and most express Cbln2. Following the fates of dI2, dI4, dI6 and dILA neurons is more problematic because these various classes of dorsal interneurons cannot be distinguished from one another by a unique marker, but only by their relative positions early in development (although they can be distinguished from Pax2-expressing v0 and v1 neurons by the expression of Evx1 and Engrail1, respectively (Burrill et al., 1997). It is generally agreed that dILA neurons migrate to the superficial dorsal horn, dI2 neurons to the deep dorsal horn, dI6 neurons to the ventral horn, but the fate of dI4 neurons appears less certain (Caspary and Anderson, 2003; Helms and Johnson; 2003; Lewis 2006). In any case, we found that Lhx1/5+ neurons and Pax2+ neurons are scattered throughout the spinal cord at St. 37, a small proportion of which express Cbln2.
Therefore, Cbln2 expression by many Lmx1b+ neurons in laminae I–IV at St. 37 is attributable to the early expression of Cbln2 by dI5 and dILB neurons. Cbln2 expression by some Pax2+ neurons in laminae I–IV at St. 37 is consistent with the early expression of Cbln2 by dILA neurons and by dI4 and/or dI6 neurons. Cbln2 expression by large neurons in lamina VI at St. 37 is consistent with the early expression of Cbln2 by Lhx9+ dI1, Islet1+ dI3, Lhx1/5+ dI2 neurons, and Lhx1/5+/Pax2+ dI4 (and/or dI6) neurons. In contrast to the high level of Cbln2 expression dorsally, most neurons in intermediate and ventral regions of the mature spinal cord do not express Cbln2, consistent with their origins from Cbln2-negative v0, v1, v2, and v3 cell populations.
Cbln2 Expression in the DRGs
In the DRGs, Cbln2 is expressed by relatively few neurons at St. 24 with that number rapidly increasing over the next day of development, as additional sensory neurons are generated and differentiate. The numerous DRG neurons expressing Cbln2 at St. 26 are spread throughout the core of the ganglia and thus would be among the older neurons in the DRG. Neurons at the periphery of the DRG and at its dorsal pole, which are generated even later and include those that are part of a secondary migration (George et al., 2007, 2010), do not express Cbln2.
In mature animals, DRG neurons are extremely diverse in their physiological properties and the types of stimuli they detect, such that more than 20 different types have been recognized (Perl, 1992). However, for some purposes including developmental studies, DRG neurons are typically grouped into a few functionally distinct types that express different neurotrophin receptors. TrkA is expressed primarily by nociceptive and thermoreceptive neurons, TrkB primarily by mechanoreceptive neurons, and TrkC primarily by proprioceptive neurons. Initially DRG neurons coexpress multiple Trks, but by St. 30, Trk expression has stabilized and individual neurons tend to express only one type of Trk receptor (Rifkin et al., 2000). Our results show that Cbln2 is expressed by nearly all TrkB+ neurons and nearly all TrkC+ neurons, but virtually no TrkA+ neurons. Thus, mechanoreceptive neurons and proprioceptive neurons express Cbln2, but nociceptive and thermoreceptive neurons do not.
Cbln2 expression by synaptically-connected neuronal populations
Our results show that Cbln2 is frequently expressed by populations of neurons that are synaptically connected. We have, in fact, been inspired by the association described here to examine the expression of Cbln2 in the chick brain, where we find that Cbln2 also frequently maps to regions in the brain that are synaptically connected, as will be reported in a separate publication (Honig, Yang, and Reiner, in preparation). With respect to the functional implications of our findings, we realize that the pattern of Cbln2 protein expression may differ from the pattern of Cbln2 mRNA expression and that it will be important to demonstrate that individual synaptically-connected neurons express Cbln2 protein. Unfortunately, an antibody suitable for carrying out such studies is not currently available.
The results presented here provide several examples of synaptically-connected Cbln2+ neuronal populations. First, in thoracic spinal cord segments, Cbln2 is heavily expressed by sympathetic preganglionic neurons in the column of Terni and by the sympathetic chain ganglion neurons to which they project.
Second, Cbln2 is expressed by TrkC+ sensory neurons and large interneurons in lamina VI. Intracellular labeling in mammals has also shown that Ia muscle spindle afferents and Ib Golgi tendon organ afferents, which both express TrkC (e.g. Hasegawa and Wang, 2008) form terminals in lamina VI (summarized in Fyffe, 1992), in addition to Ia afferents synapsing on motoneurons in the lateral motor column. In the chick, DiI labeling has revealed that muscle nerve afferents arborize extensively in lamina VI (Eide and Glover, 1997) and immunolabeling has shown that TrkC+ sensory fibers terminate in this general region (Oakley et al., 1997). Here, by combining labeling dorsal roots and immunofluorescent staining for TrkC and for laminar markers, we have demonstrated that TrkC+ sensory afferents arborize in lamina VI, in the vicinity of the large neurons that express Cbln2. Interestingly, many of the large neurons in lamina VI are spinocerebellar tract neurons (Yamamoto et al., 2000), which are known to receive input from muscle receptors (Kuno et al., 1973).
Third, Cbln2 is expressed by TrkB+ and TrkC+ neurons in the DRGs, and by interneurons in lamina II and lamina III, and to a lesser extent in the medial part of lamina IV. To our surprise, there is little published data on the projections of TrkB+ and TrkC+ sensory axons in the dorsal spinal cord. When we examined these projections ourselves, we found that TrkB+ sensory axons arborize in lamina II, lamina III and medial lamina IV and TrkC+ sensory axons in lamina II and medial lamina IV. Our results differ somewhat from those of Rifkin et al. (2000), who found that TrkB+ and TrkC+ afferents terminate primarily in lamina II and lamina III, possibly because we examined these projections in slightly older embryos and in conjunction with markers to distinguish among laminae. Our results are consistent with retrograde labeling of myelinated cutaneous nerve axons in chicks (Woodbury and Scott, 1991; see also Eide and Glover, 1997) and intracellular labeling of myelinated mechanoreceptive afferents in mammals (Fyffe, 1992), which demonstrate that myelinated cutaneous nerve axons primarily project into laminae III and IV. In addition, some mechanoreceptive cutaneous afferents arborize in lamina II (Rethelyi et al., 1989) and some interneurons in lamina II respond to mechanical stimulation of the skin (Rethelyi et al., 1989; Woodbury, 1992). While it is frequently stated that mechanoreceptive cutaneous afferents express TrkB, some TrkC+ sensory neurons also innervate the skin (in chick, Oakley et al., 1995; in mouse, Airaksinen et al., 1996; Hasegawa and Wang, 2008), and they would be expected to project to the dorsal horn. Indeed, in accord with this, we found a substantial TrkC projection to lamina II and medial lamina IV. It seems likely that the TrkC projection to dorsal laminae may often be overlooked due to the frequent focus on projections from muscle spindle and Golgi tendon organ afferents to the intermediate and ventral spinal cord (Oakley et al., 1997; Yoshida et al., 2006; but see Nakamura et al., 2008). Thus, Cbln2 is expressed by TrkB+ sensory neurons and their targets in lamina II, lamina III and medial lamina IV, and by TrkC+ sensory neurons and their targets in lamina II and medial lamina IV. Notably, Cbln2 is expressed at a much lower level in lamina I and lateral lamina IV where TrkA+ afferents, which are Cbln2-negative, project nearly exclusively.
We cannot exclude the possibility that there are additional and/or alternative explanations for the differential expression of Cbln2 within the dorsal horn. Certainly, laminae I–IV contain neurons with a variety of somatodendritic morphologies, electrophysiological properties, projections to different parts of the spinal cord and to the brain, and inputs from sources in addition to the DRGs. For example, spinocervical neurons and neurons with axons ascending in the dorsal columns are present in lamina IV, and to a lesser extent in lamina III. Similarly, various descending systems terminate predominantly in two or three laminae. However, in these and most other respects, lamina I and lamina II are similar to one other and lamina III and lamina IV are similar to one another (Brown, 1981). In contrast, we found that lamina II and lamina III share nearly identical levels of Cbln2 expression and the characteristic most shared by lamina II and lamina III is that they receive input from TrkB+ and TrkC+ sensory afferents. In addition, Cbln2 is more highly expressed by the medial part of lamina IV, which receives input from TrkB+ and TrkC+ sensory afferents, than by the lateral part of lamina IV, which lacks substantial TrkB and TrkC input.
It should be acknowledged that some (approximately one-fourth) of the neurons in lamina I and the lateral part of lamina IV express Cbln2. These regions receive some TrkB and TrkC input, albeit at much lower levels than they receive TrkA input. This, together with other aspects of the results, raises the question of whether the tendency for Cbln2 to be expressed by neuronal populations that are synaptically connected extends to the level of individual neurons. Laminae I–IV of the mature dorsal horn are populated by glutamatergic excitatory and GABAergic inhibitory interneurons, which differentiate from Lmx1b+ and Pax2+ neurons, respectively (Cheng et al., 2004, 2005). We found that Cbln2 is expressed by both Lmx1b+ neurons and Pax2+ neurons, but that the percentage of Lmx1b+ neurons expressing Cbln2 is more than twice that for Pax2+ neurons. While it is widely acknowledged that sensory afferents terminate on the glutamatergic interneurons (Cheng et al., 2004), sensory afferents also form synapses with GABAergic interneurons (Bernardi et al., 1995; Grudt and Perl 2002; Lu and Perl 2005; Braz and Basbaum, 2009). However, it is not known, for example, if sensory afferents synapse preferentially with the excitatory interneurons, or if all the excitatory interneurons receive monosynaptic sensory input. Moreover, different types of afferents exhibit different patterns of terminal arborizations (Fyffe, 1992), and so would be expected to exhibit different patterns of synaptic connections, but these have not yet been described in detail. Thus, given that much basic information about circuitry in the dorsal horn is currently lacking, whether the differential expression of Cbln2 reflects the underlying circuitry, or whether there are other characteristics that distinguish interneurons expressing Cbln2 from those that do not, cannot yet be resolved. Nonetheless, one simple possibility is that Cbln2+ DRG neurons (TrkB+ and TrkC+) synapse with Cbln2+ neurons in laminae I–IV, and the differential expression of Cbln2 in the dorsal horn reflects the preferential targeting by TrkB+ and TrkC+ afferents.
Role of Cblns in synapse formation and maintenance
Current understanding of the function of any of the Cblns is based almost exclusively on experiments addressing the role of Cbln1 in the cerebellum. Cbln1 is synthesized by granule cells and released from their terminals onto the dendritic spines of Purkinje cells (Hirai et al., 2005). Cbln1 knockout (KO) mice are ataxic and, while Purkinje cell dendritic spines themselves appear normal, there is a severe reduction in the number of PF synapses on those spines, and a corresponding deficit in synaptic transmission. Importantly, recombinant Cbln1 has been shown to rescue the effect of the loss of Cbln1 by inducing PF synapses in cultures of dissociated cells, in slices and in the KO animal itself (Ito-Ishida et al., 2008). Expressing Cbln1 ectopically in Purkinje cells also has the effect of rescuing locomotor deficits in Cbln1-null mice (Wei et al., 2009). Interestingly, the expression of Cbln1 mRNA and protein is influenced by electrical activity, with Cbln1 levels decreasing when granule cells are depolarized or when voltage-gated calcium channels are blocked, thereby providing a means by which these synapses may maintain homeostasis in normal animals (Iijima et al., 2009).
Although it is clear that Cbln1 plays an important role at the synapse, many questions about how Cbln1 carries out this function remain unanswered. After its release from PF terminals, Cbln1 is internalized by the dendritic spines of Purkinje cells and becomes localized within late endosomes and lysosomes. One possibility is that the internalized Cbln1 subsequently initiates a signal transduction cascade within endosomes, analogous to the means by which neurotrophins exert long-range effects (Wei et al., 2009). It has also recently been shown that Cbln1 hexamers bind to the postsynaptic surface of Purkinje cells (Miura et al., 2009; Matsuda et al., 2009), suggesting the presence of a specific membrane-bound receptor. Mice lacking GluRdelta2, an orphan glutamate receptor expressed on Purkinje cells, are characterized by the same deficits as Cbln1 KO mice (Hirai et al., 2005), consistent with the possibility that GluRdelta2 is that receptor. However, the absence of GluRdelta2 in numerous other brain regions where Cbln1 is present supports the alternative view that Cbln1 and GluRdelta2 may instead be involved in a common signaling pathway and another Cbln1 receptor must exist (e.g. Hirai et al., 2005; Wei et al., 2009). Regardless, for any of these possibilities, the mechanisms underlying Cbln1 action appear to be different from those elucidated for other molecules contributing to synapse formation, for example neuroligans, which often involve adhesive interactions between pre- and post-synaptic cell membranes (McAllister, 2007; Barrow et al., 2009; Lucido et al., 2009).
To the best of our knowledge, the function of Cbln2 has not been examined. However, the amino acid sequence of Cbln2 is very similar to that of Cbln1 (for the mouse homologues, 70% for the whole protein, 87% for the C1q domain), and so Cbln2 may also function in synapse formation and maintenance. Cbln2 is abundant in numerous brain regions (Wada and Ohtani, 1991; Miura et al., 2006), and as we have shown here, in the DRGs and spinal cord. Further, it is secreted as a glycosylated protein from heterologous cells (Iijima et al., 2007; J. Morgan, personal communication), and so it is likely that it will be secreted from neurons in vivo as well. Moreover, many sensory and spinal cord neurons express Cbln2 by St. 26, long before sensory axons form appropriate projections within the dorsal horn (Davis et al., 1989; Eide and Glover, 1995), consistent with the possibility that Cbln2 participates in the formation of synapses, as well as in their maintenance. Determining whether Cbln2 indeed plays such roles during development and in mature animals will be facilitated by the analysis of Cbln2 KO mice.
All four Cblns can form heteromers in heterologous cell cultures. Cbln1 and Cbln3 also form heteromers in cultured granule cells and, most importantly, in vivo. Moreover, Cbln1/3 heteromers appear to function differently than do either Cbln1 homomers or Cbln3 homomers. In Cbln1 KO mice, Cbln3 gene expression appears normal but the Cbln3 protein is retained internally and most of it is ultimately degraded (Bao et al., 2006; Iijima et al., 2007; Wei et al., 2007). Cbln3 KO mice express higher than normal levels of Cbln1 protein and are phenotypically normal, whereas Cbln1/3 double KOs show deficits in PF-Purkinje cell synapses, thereby phenocopying the Cbln1 single KO (Bao et al., 2006). Thus, Cbln1 and Cbln3 interact in complex ways, with each influencing the other’s degradation and secretion. Interestingly, the pattern of Cbln2 expression often overlaps that of Cbln1 in the mouse brain (Miura et al., 2006). If individual neurons coexpress Cbln1 and Cbln2, these two Cblns may form heteromers in vivo, and in turn, this may have important functional implications.
We find it extremely intriguing that Cbln2 mRNA is expressed by synaptically connected populations of neurons. This observation raises the possibility that Cbln2 participates in bidirectional signaling between pre- and postsynaptic neurons and suggests Cbln2 may act somewhat differently than Cbln1 has been shown to in the cerebellum. Interestingly, however, a recent report demonstrating that Cbln1 can travel in both directions across PF-Purkinje cell synapses in vivo (Wei et al., 2009), suggests that Cbln1 may function not only in the postsynaptic neuron, but also in the presynaptic neuron. It will therefore be important to elucidate Cbln2’s function at the cellular level and its effects on synapse formation and maintenance.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. James Morgan for sharing his knowledge of cerebellins with us and for helpful comments on the manuscript. We are grateful to Dr. Thomas Müller and Dr. Carmen Birchmeier; Max-Delbrück-Centrum for Molecular Medicine. Berlin, Germany, for the Lmx1b antibody and to Dr. Frances Lefcort, Montana State University, for the Trk antibodies. Antibodies against Islet1 (39.4D5), Lhx1/5 (4F2), Engrail1 (4G11), and Evx1/2 (99.1-3A2) were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. The project described was supported by NIH Grant Number NS34404 to MGH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
LITERATURE CITED
- Airaksinen MS, Koltzenburg M, Lewin GR, Masu Y, Helbig C, Wolf E, Brem G, Toyka KV, Thoenen H, Meyer M. Specific subtypes of cutaneous mechanoreceptors require Neurotrophin-3 following peripheral target innervation. Neuron. 1996;16:287–295. doi: 10.1016/s0896-6273(00)80047-1. [DOI] [PubMed] [Google Scholar]
- Bao D, Pang Z, Morgan JI. The structure and proteolytic processing of Cbln1 complexes. J Neurochem. 2005;95:618–629. doi: 10.1111/j.1471-4159.2005.03385.x. [DOI] [PubMed] [Google Scholar]
- Bao D, Pang Z, Morgan MA, Parris J, Rong Y, Li L, Morgan JI. Cbln1 is essential for interaction-dependent secretion of Cbln3. Mol Cell Biol. 2006;26:9327–9337. doi: 10.1128/MCB.01161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrow SL, Constable JR, Clark E, El-Sabeawy F, McAllister AK, Washbourne P. Neuroligin1: a cell adhesion molecule that recruits PSD-95 and NMDA receptors by distinct mechanisms during synaptogenesis. Neural Dev. 2009;4:17. doi: 10.1186/1749-8104-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on MATH1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Bernardi PS, Valtschanoff JG, Weinberg RJ, Schmidt HH, Rustioni A. Synaptic interactions between primary afferent terminals and GABA and nitric oxide-synthesizing neurons in superficial laminae of the rat spinal cord. J Neurosci. 1995;15:1363–1371. doi: 10.1523/JNEUROSCI.15-02-01363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braz JM, Basbaum AI. Triggering genetically-expressed transneuronal tracers by peripheral axotomy reveals convergent and segregated sensory neuron-spinal cord connectivity. Neuroscience. 2009;163:1220–1232. doi: 10.1016/j.neuroscience.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman R, Martin AH. A cytoarchitectonic study of the spinal cord of the domestic fowl Gallus gallus domesticus. I. Brachial region. Brain Res. 1973;56:43–62. doi: 10.1016/0006-8993(73)90326-0. [DOI] [PubMed] [Google Scholar]
- Brown AG. Organization in the spinal cord, the anatomy and physiology of identified neurons. Berlin: Springer-Verlag; 1981. [Google Scholar]
- Burrill JD, Moran L, Goulding MD, Saueressig H. PAX2 is expressed in multiple spinal cord interneurons, including a population of EN1+ interneurons that require PAX6 for their development. Development. 1997;124:4493–4503. doi: 10.1242/dev.124.22.4493. [DOI] [PubMed] [Google Scholar]
- Carr VM, Simpson SB., Jr Proliferative and degenerative events in the early development of chick dorsal root ganglia. I. Normal development. J Comp Neurol. 1978;182:727–739. doi: 10.1002/cne.901820410. [DOI] [PubMed] [Google Scholar]
- Caspary T, Anderson KV. Patterning cell types in the dorsal spinal cord: what the mouse mutants say. Nat Rev Neurosci. 2003;4:289–297. doi: 10.1038/nrn1073. [DOI] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, Chen CL, Busslinger M, Goulding M, Onimaru H, Ma Q. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nature Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Millen KJ. Control of roof plate development and signaling by Lmx1b in the caudal vertebrate CNS. J Neurosci. 2004;24:5694–5703. doi: 10.1523/JNEUROSCI.0758-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BM, Frank E, Johnson FA, Scott SA. Development of central projections of lumbosacral sensory neurons in the chick. J Comp Neurol. 1989;279:556–566. doi: 10.1002/cne.902790405. [DOI] [PubMed] [Google Scholar]
- Davis CA, Holmyard DP, Millen KJ, Joyner AL. Examining pattern formation in mouse, chicken and frog embryos with an En-specific antiserum. Development. 1991;111:287–298. doi: 10.1242/dev.111.2.287. [DOI] [PubMed] [Google Scholar]
- Ding YQ, Yin J, Kania A, Zhao ZQ, Johnson RL, Chen ZF. Lmx1b controls the differentiation and migration of the superficial dorsal horn neurons of the spinal cord. Development. 2004;131:3693–3703. doi: 10.1242/dev.01250. [DOI] [PubMed] [Google Scholar]
- Dressler GR, Douglass EC. Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc Natl Acad Sci USA. 1992;89:1179–1183. doi: 10.1073/pnas.89.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubus P, Faucheux B, Boissière F, Groppi A, Vital C, Vital A, Agid Y, Hirsch EC, Merlio JP. Expression of Trk isoforms in brain regions and in the striatum of patients with Alzheimer's disease. Exp Neurol. 2000;165:285–294. doi: 10.1006/exnr.2000.7447. [DOI] [PubMed] [Google Scholar]
- Eide AL, Glover JC. Development of the longitudinal projection patterns of lumbar primary sensory afferents in the chicken embryo. J Comp Neurol. 1995;353:247–259. doi: 10.1002/cne.903530207. [DOI] [PubMed] [Google Scholar]
- Eide AL, Glover JC. Developmental dynamics of functionally specific primary sensory afferent projections in the chicken embryo. Anat Embryol (Berl) 1997;195:237–250. doi: 10.1007/s004290050043. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal cell fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Fyffe REW. Laminar organization of primary afferent terminations in the mammalian spinal cord. In: Scott SA, editor. Sensory neurons: diversity development and plasticity. New York: Oxford Univ Press; 1992. pp. 131–139. [Google Scholar]
- Garner AS, Large TH. Isoforms of the avian TrkC receptor: a novel kinase insertion dissociates transformation and process outgrowth from survival. Neuron. 1994;13:457–472. doi: 10.1016/0896-6273(94)90360-3. [DOI] [PubMed] [Google Scholar]
- Garner AS, Menegay HJ, Boeshore KL, Xie XY, Voci JM, Johnson JE, Large TH. Expression of TrkB receptor isoforms in the developing avian visual system. J Neurosci. 1996;16:1740–1752. doi: 10.1523/JNEUROSCI.16-05-01740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George L, Chaverra M, Todd V, Lansford R, Lefcort F. Nociceptive sensory neurons derive from contralaterally migrating, fate-restricted neural crest cells. Nat Neurosci. 2007;10:1287–1293. doi: 10.1038/nn1962. [DOI] [PubMed] [Google Scholar]
- George L, Kasemeier-Kulesa J, Nelson BR, Koyano-Nakagawa N, Lefcort F. Patterned assembly and neurogenesis in the chick dorsal root ganglion. J Comp Neurol. 2010;518:405–422. doi: 10.1002/cne.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JC, Petursdottir G, Jansen JK. Fluorescent dextran-amines used as axonal tracers in the nervous system of the chicken embryo. J Neurosci Methods. 1986;18:243–254. doi: 10.1016/0165-0270(86)90011-7. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/s0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Grudt TJ, Perl ER. Correlations between neuronal morphology and electrophysiological features in the rodent superficial dorsal horn. J Physiology. 2002;540:189–207. doi: 10.1113/jphysiol.2001.012890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W, Condic ML. Characterization of Netrin-1, Neogenin and cUNC-5H3 expression during chick dorsal root ganglia development. Gene Expr Patterns. 2003;3:369–373. doi: 10.1016/s1567-133x(03)00007-3. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Levi-Montalcini R. Proliferation, differentiation and degeneration in the spinal ganglia of the chick embryo under normal and experimental conditions. J Exp Zool. 1949;111:457–502. doi: 10.1002/jez.1401110308. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. J Morphol. 1951;88:49–92. [PubMed] [Google Scholar]
- Hasegawa H, Wang F. Visualizing mechanosensory endings of TrkC-expressing neurons in HS3ST-2-hPLAP mice. J Comp Neurol. 2008;511:543–556. doi: 10.1002/cne.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms AW, Johnson JE. Specification of dorsal spinal cord interneurons. Current Opinion Neurobiol. 2003;13:42–49. doi: 10.1016/s0959-4388(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Hirai H, Pang Z, Bao D, Miyazaki T, Li L, Miura E, Parris J, Rong Y, Watanabe M, Yuzaki M, Morgan JI. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nature Neurosci. 2005;8:1534–1541. doi: 10.1038/nn1576. [DOI] [PubMed] [Google Scholar]
- Hollyday M, Hamburger V. An autoradiographic study of the formation of the lateral motor column in the chick embryo. Brain Res. 1977;132:197–208. doi: 10.1016/0006-8993(77)90416-4. [DOI] [PubMed] [Google Scholar]
- Honig MG. The development of sensory projection patterns in embryonic chick hindlimb. J Physiol. 1982;330:175–202. doi: 10.1113/jphysiol.1982.sp014336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honig MG, Rutishauser US. Changes in the segmental pattern of sensory neuron projections in the chick hindlimb under conditions of altered cell adhesion molecule function. Dev Biol. 1996;175:325–337. doi: 10.1006/dbio.1996.0118. [DOI] [PubMed] [Google Scholar]
- Honig MG, Frase PA, Camilli SJ. The spatial relationships among cutaneous, muscle sensory and motoneuron axons during development of the chick hindlimb. Development. 1998;125:995–1004. doi: 10.1242/dev.125.6.995. [DOI] [PubMed] [Google Scholar]
- Honig MG, Camilli SJ, Surineni KM, Knight BK, Hardin HM. The contributions of BMP4, positive guidance cues, and repulsive molecules to cutaneous nerve formation in the chick hindlimb. Dev Biol. 2005;282:257–273. doi: 10.1016/j.ydbio.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Iijima T, Miura E, Matsuda K, Kamekawa Y, Watanabe M, Yuzaki M. Characterization of a transneuronal cytokine family Cbln-regulation of secretion by heteromeric assembly. Eur J Neurosci. 2007;25:1049–1057. doi: 10.1111/j.1460-9568.2007.05361.x. [DOI] [PubMed] [Google Scholar]
- Iijima Y, Emi K, Yuzaki M. Activity-dependent repression of Cbln1 expression: mechanism for developmental and homeostatic regulation of synapses in the cerebellum. J Neurosci. 2009;29:5425–5434. doi: 10.1523/JNEUROSCI.4473-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Ishida A, Miura E, Emi K, Matsuda K, Iijima T, Kondo T, Kohda K, Watanabe M, Yuzaki M. Cbln1 regulates rapid formation and maintenance of excitatory synapses in mature cerebellar Purkinje cells in vitro and in vivo. J Neurosci. 2008;28:5920–5930. doi: 10.1523/JNEUROSCI.1030-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno M, Musoz-Martinez EJ, Randic M. Sensory inputs to neurons in Clarke’s column from muscle, cutaneous and joint receptors. J Physiol. 1973;228:327–342. doi: 10.1113/jphysiol.1973.sp010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lance-Jones C, Landmesser LT. Pathway selection by chick lumbosacral motoneurons during normal development. Pro Roy Soc London. 1981;214:1–18. doi: 10.1098/rspb.1981.0079. [DOI] [PubMed] [Google Scholar]
- Landmesser LT. The development of motor projection patterns in the chick hind limb. J Physiol. 1978;284:391–414. doi: 10.1113/jphysiol.1978.sp012546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser LT, Honig MG. Altered sensory projections in the chick hindlimb following the early removal of motoneurons. Dev Biol. 1986;118:511–531. doi: 10.1016/0012-1606(86)90023-0. [DOI] [PubMed] [Google Scholar]
- Lawoko-Kerali G, Rivolta MN, Lawlor P, Cacciabue-Rivolta DI, Langton-Hewer C, van Doorninck JH, Holley MC. GATA3 and NeuroD distinguish auditory and vestibular neurons during development of the mammalian inner ear. Mechanisms Devel. 2004;121:287–299. doi: 10.1016/j.mod.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Lee MK, Tuttle JB, Rebhun LI, Cleveland DW, Frankfurter A. The expression and posttranslational modification of a neuron-specific beta-tubulin isotype during chick embryogenesis. Cell Motil Cytoskeleton. 1990;17:118–132. doi: 10.1002/cm.970170207. [DOI] [PubMed] [Google Scholar]
- Lee KJ, Mendelsohn M, Jessell TM. Neuronal patterning by BMPs: a requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev. 1998;12:3394–3407. doi: 10.1101/gad.12.21.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefcort F, Clary DO, Rusoff AC, Reichardt LF. Inhibition of the NT-3 receptor TrkC, early in chick embryogenesis, results in severe reductions in multiple neuronal subpopulations in the dorsal root ganglia. J Neurosci. 1996;16:3704–3713. doi: 10.1523/JNEUROSCI.16-11-03704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KE. How do genes regulate simple behaviors? Understanding how different neurons in the vertebrate spinal cord are genetically specified. Phil Trans R Soc B. 2006;361:45–66. doi: 10.1098/rstb.2005.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, Jr, Tremml G, Jessell TM. A role for the roof plate and its resident TGFbeta-related proteins in neuronal patterning in the dorsal spinal cord. Cell. 1997;91:127–138. doi: 10.1016/s0092-8674(01)80015-5. [DOI] [PubMed] [Google Scholar]
- Lin JH, Saito T, Anderson DJ, Lance-Jones C, Jessell TM, Arber S. Functionally related motor neuron pool and muscle sensory afferent subtypes defined by coordinate ETS gene expression. Cell. 1998;95:393–407. doi: 10.1016/s0092-8674(00)81770-5. [DOI] [PubMed] [Google Scholar]
- Lind D, Franken S, Kappler J, Jankowski J, Schilling K. Characterization of the neuronal marker NeuN as a multiply phosphorylated antigen with discrete subcellular localization. J Neurosci Res. 2005;79:295–302. doi: 10.1002/jnr.20354. [DOI] [PubMed] [Google Scholar]
- Lu Y, Perl ER. Modular organization of excitatory circuits between neurons of the spinal superficial dorsal horn (laminae I and II) J Neurosci. 2005;25:3900–3907. doi: 10.1523/JNEUROSCI.0102-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucido AL, Suarez Sanchez F, Thostrup P, Kwiatkowski AV, Leal-Ortiz S, Gopalakrishnan G, Liazoghli D, Belkaid W, Lennox RB, Grutter P, Garner CC, Colman DR. Rapid assembly of functional presynaptic boutons triggered by adhesive contacts. J Neurosci. 2009;29:12449–12466. doi: 10.1523/JNEUROSCI.1381-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AH. A cytoarchitectonic scheme for the spinal cord of the domestic fowl, Gallus gallus domesticus: lumbar region. Acta Morphol Neerl Scand. 1979;17:105–117. [PubMed] [Google Scholar]
- Marusich MF, Furneaux HM, Henion PD, Weston JA. Hu neuronal proteins are expressed in proliferating neurogenic cells. J Neurobiol. 1994;25:143–155. doi: 10.1002/neu.480250206. [DOI] [PubMed] [Google Scholar]
- Matise MP, Joyner AL. Expression patterns of developmental control genes in normal and Engrailed-1 mutant mouse spinal cord reveal early diversity in developing interneurons. J Neurosci. 1997;17:7805–7816. doi: 10.1523/JNEUROSCI.17-20-07805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Kondo T, Iijima T, Matsuda S, Watanabe M, Yuzaki M. Cbln1 binds to specific postsynaptic sites at parallel fiber-Purkinje cell synapses in the cerebellum. Eur J Neurosci. 2009;29:693–706. doi: 10.1111/j.1460-9568.2009.06639.x. [DOI] [PubMed] [Google Scholar]
- McAllister AK. Dynamic aspects of CNS synapse formation. Annu Rev Neurosci. 2007;30:425–450. doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesegaes GR, Klisch TJ, Thaller C, Ahmad KA, Atkinson RC, Zoghbi HY. Identification and subclassification of new Atoh1 derived cell populations during mouse spinal cord development. Dev Biol. 2009;327:339–351. doi: 10.1016/j.ydbio.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura E, Iijima T, Yuzaki M, Watanabe M. Distinct expression of Cbln family mRNAs in developing and adult mouse brains. Eur J Neurosci. 2006;24:750–760. doi: 10.1111/j.1460-9568.2006.04950.x. [DOI] [PubMed] [Google Scholar]
- Miura E, Matsuda K, Morgan JI, Yuzaki M, Watanabe M. Cbln1 accumulates and colocalizes with Cbln3 and GluRdelta2 at parallel fiber-Purkinje cell synapses in the mouse cerebellum. Eur J Neurosci. 2009;29:693–706. doi: 10.1111/j.1460-9568.2009.06632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RJ, Buck CR, Smith AM. NeuN, a neuronal specific nuclear protein in vertebrates. Development. 1992;116:201–211. doi: 10.1242/dev.116.1.201. [DOI] [PubMed] [Google Scholar]
- Muller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor Lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/s0896-6273(02)00689-x. [DOI] [PubMed] [Google Scholar]
- Muller T, Anlag K, Wildner H, Britsch S, Treier M, Birchmeier C. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes Devel. 2005;19:733–743. doi: 10.1101/gad.326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz LM, Zayachkivsky A, Kunz RB, Hunt JM, Wang G, Scott SA. Ephrin-A5 inhibits growth of embryonic sensory neurons. Dev Biol. 2005;283:397–408. doi: 10.1016/j.ydbio.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Senzaki K, Yoshikawa M, Nishimura M, Inoue K, Ito Y, Ozaki S, Shiga T. Dynamic regulation of the expression of neurotrophin receptors by Runx3. Development. 2008;135:1703–1711. doi: 10.1242/dev.015248. [DOI] [PubMed] [Google Scholar]
- Oakley RA, Garner AS, Large TH, Frank E. Muscle sensory neurons require neurotrophin-3 from peripheral tissues during the period of normal cell death. Development. 1995;121:1341–1350. doi: 10.1242/dev.121.5.1341. [DOI] [PubMed] [Google Scholar]
- Oakley RA, Lefcort FB, Clary DO, Reichardt LF, Prevette D, Oppenheim RW, Frank E. Neurotrophin-3 promotes the differentiation of muscle spindle afferents in the absence of peripheral targets. J Neurosci. 1997;17:4262–4274. doi: 10.1523/JNEUROSCI.17-11-04262.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Zuo J, Morgan JI. Cbln3, a novel member of the precerebellin family that binds specifically to Cbln1. J Neurosci. 2000;20:6333–6339. doi: 10.1523/JNEUROSCI.20-17-06333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl ER. Function of dorsal ganglion neurons: an overview. In: Scott SA, editor. Sensory neurons: diversity development and plasticity. New York: Oxford Univ Press; 1992. pp. 3–23. [Google Scholar]
- Pierani A, Brenner-Morton S, Chiang C, Jessell TM. A Sonic hedgehog-independent, retinoid-activated pathway of neurogenesis in the ventral spinal cord. Cell. 1999;97:903–915. doi: 10.1016/s0092-8674(00)80802-8. [DOI] [PubMed] [Google Scholar]
- Prasad A, Hollyday M. Development and migration of avian sympathetic preganglionic neurons. J Comp Neurol. 1991;307:237–258. doi: 10.1002/cne.903070207. [DOI] [PubMed] [Google Scholar]
- Qian Y, Shirasawa S, Chen CL, Cheng L, Ma Q. Proper development of relay somatic sensory neurons and D2/D4 interneurons requires homeobox genes Rnx/Tlx-3 and Tlx-1. Genes Dev. 2002;16:1220–1233. doi: 10.1101/gad.982802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethelyi M, Light AR, Perl ER. Synaptic ultrastructure of functionally and morphologically characterized neurons of the superficial spinal dorsal horn of cat. J Neurosci. 1989;9:1848–l863. doi: 10.1523/JNEUROSCI.09-06-01846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin JT, Todd VJ, Anderson LW, Lefcort F. Dynamic expression of neurotrophin receptors during sensory neuron genesis and differentiation. Dev Biol. 2000;227:465–480. doi: 10.1006/dbio.2000.9841. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Simmons DM, Arriza JL, Swanson LW. A complete protocol for in situ hybridization of messenger RNAs in brain and other tissues with radio-labeled single-stranded RNA probes. J Histotechnol. 1989;12:169–181. [Google Scholar]
- Slemmon JR, Russell Blacher R, Danho W, Hempstead JL, Morgan JI. Isolation and sequencing of two cerebellum-specific peptides. Proc Natl Acad Sci USA. 1984;81:6866–6870. doi: 10.1073/pnas.81.21.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern CD. Detection of multiple gene products simultaneously by in situ hybridization and immunohistochemistry in whole mounts of avian embryos. Curr Top Dev Biol. 1998;36:223–243. doi: 10.1016/s0070-2153(08)60505-0. [DOI] [PubMed] [Google Scholar]
- Stoeckli ET, Landmesser LT. Axonin-1, Nr-CAM, and Ng-CAM play different roles in the in vivo guidance of chick commissural neurons. Neuron. 1995;14:1165–1179. doi: 10.1016/0896-6273(95)90264-3. [DOI] [PubMed] [Google Scholar]
- Tang YT, Hu T, Arterburn M, Boyle B, Bright JM, Palencia S, Emtage PC, Funk WD. The complete complement of C1q-domain-containing proteins in Homo sapiens. Genomics. 2005;86:100–111. doi: 10.1016/j.ygeno.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Koo SJ, Kania A, Lettieri K, Andrews S, Cox C, Jessell TM, Pfaff SL. A postmitotic role for Isl-class LIM homeodomain proteins in the assignment of visceral spinal motor neuron identity. Neuron. 2004;41:337–350. doi: 10.1016/s0896-6273(04)00011-x. [DOI] [PubMed] [Google Scholar]
- Tosney KW, Landmesser LT. Development of the major pathways for neurite outgrowth in the chick hindlimb. Dev Biol. 1985;109:193–214. doi: 10.1016/0012-1606(85)90360-4. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Urade Y, Oberdick J, Molinar-Rode R, Morgan JI. Precerebellin is a cerebellum-specific protein with similarity to the globular domain of complement C1q B chain. Proc Natl Acad Sci USA. 1991;88:1069–1073. doi: 10.1073/pnas.88.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada C, Ohtani H. Molecular cloning of rat cerebellin-like protein cDNA which encodes a novel membrane-associated glycoprotein. Mol Brain Res. 1991;9:71–77. doi: 10.1016/0169-328x(91)90131-g. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Weston JA. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development. 1997;124:3449–3460. doi: 10.1242/dev.124.17.3449. [DOI] [PubMed] [Google Scholar]
- Wei P, Smeyne RJ, Bao D, Parris J, Morgan JI. Mapping of Cbln1-like immunoreactivity in adult and developing mouse brain and its localization to the endolysosomal compartment of neurons. Eur J Neurosci. 2007;26:2962–2978. doi: 10.1111/j.1460-9568.2007.05913.x. [DOI] [PubMed] [Google Scholar]
- Wei P, Rong Y, Li L, Bao D, Morgan JI. Characterization of trans-neuronal trafficking of Cbln1. Mol Cell Neurosci. 2009;41:258–273. doi: 10.1016/j.mcn.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildner H, Muller T, Cho SH, Brohl D, Cepko CL, Guillemot F, Birchmeier C. dILA neurons in the dorsal spinal cord are the product of terminal and non-terminal asymmetric progenitor cell divisions, and require Mash1 for their development. Development. 2006;133:2105–2113. doi: 10.1242/dev.02345. [DOI] [PubMed] [Google Scholar]
- Woodbury CJ, Scott SA. Somatotopic organization of hindlimb skin sensory inputs to the dorsal horn of hatchling chicks (Gallus g. domesticus) J Comp Neurol. 1991;314:237–256. doi: 10.1002/cne.903140204. [DOI] [PubMed] [Google Scholar]
- Woodbury CJ. Physiological studies of cutaneous inputs to dorsal horn laminae I-IV of adult chickens. J Neurophys. 1992;67:241–254. doi: 10.1152/jn.1992.67.2.241. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Kitagawa H, Imagawa T, Uehara M. The organization of the spinocerebellar tract neurons in the chicken. Brain Res Bulletin. 2000;52:537–546. doi: 10.1016/s0361-9230(00)00296-3. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Han B, Mendelsohn M, Jessell TM. PlexinA1 signaling directs the segregation of proprioceptive sensory axons in the developing spinal cord. Neuron. 2006;52:775–788. doi: 10.1016/j.neuron.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzaki M. Cbln and C1q family proteins - new transneuronal cytokines. Cell Mol Life Sci. 2008;65:1698–1705. doi: 10.1007/s00018-008-7550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.