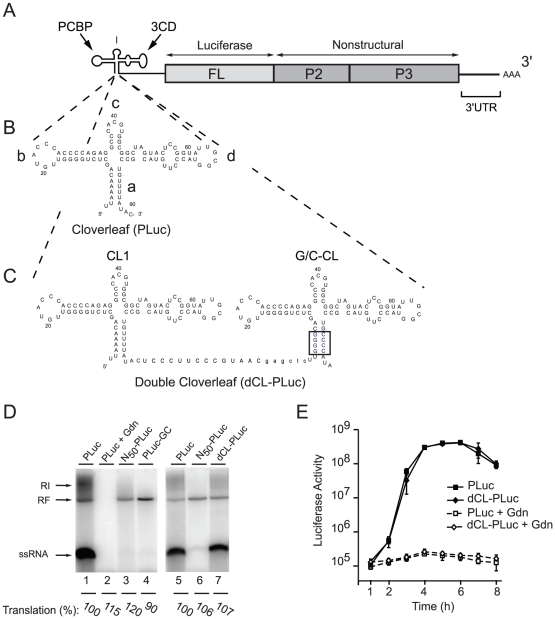
Figure 1. Double cloverleaf replicons.
(A) Schematic representation of a poliovirus-luciferase replicon. (B and C) Secondary structure of the PLuc cloverleaf and the two tandem cloverleaves in dCL-PLuc with wild-type sequences in CL1 and four G-C pairs in “stem a” of the downstream cloverleaf G/C-CL (highlighted in blue). (D) RNA replication in a cell-free system. RNA transcripts either Pluc (lanes 1, 2+5), N50-Pluc (50 non-polio nucleotides at the 5′-end of the viral RNA) (lanes 3+6), Pluc-GC (lane 4), or dCL-Pluc (lane 7) were used to program HeLa cell S10 extract. After 4 hours of incubation at 30°C, translation levels were measured as luciferase activity (arbitrary units [AU]), and pre-initiation complexes were isolated by centrifugation in the presence of 2 mM guanidinium hydrochloride (Gdn). RNA synthesis was initiated by addition of NTP and monitored by [α32P]UTP incorporation for 2 hr. The RNA synthesized was analyzed using native agarose gels. (E) Replication of poliovirus replicon in intact HeLa S3 cells. RNA transcripts (PLuc or dCL-PLuc) were transfected into HeLa S3 cells, and luciferase activity [AU] corresponding to 2.5×105 cells was measured every hour for 8h. The cells were incubated either in the presence (dashed lines) or absence (solid lines) of 2 mM Gdn. Each measurement was carried out in triplicate; standard deviations are indicated by vertical bars.