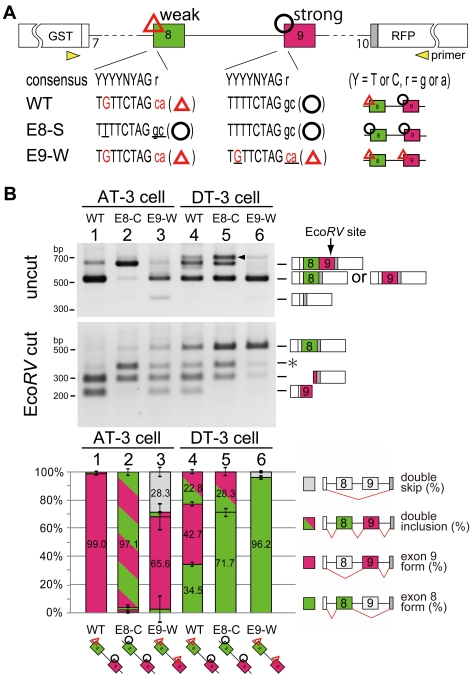
Figure 2. Unbalanced sequence of 3′ splice sites is essential for mutually exclusive exon selection.
(A) Scheme for 3' splice site mutation on exons 8 and 9. Uppercase letter is intron and lowercase is exon sequence. Red characters indicate mismatches from the conserved consensus sequence in the 3' splice site and poly-pyrimidine moiety, and underline indicate mutated sequence. The yellow arrows with “primer” represent the positions amplified in RT-PCR. (B) RT-PCR from AT-3 and DT-3 cells transfected the indicated vectors. Splice products were digested with EcoRV, which uniquely cuts the PCR product containing exon 9. Each band was identified and indicated with the scheme of splice products. Arrowheads indicate nonspecific PCR products, which was confirmed by sequencing. The asterisk indicates the splice product came from double inclusion of exon 8 and exon 9. The bar graph shows the amount of each splicing product, and is based on calculations from three independent experiments; the mean value for each splice product is show in the respective column with an error bar showing the SD (standard error).