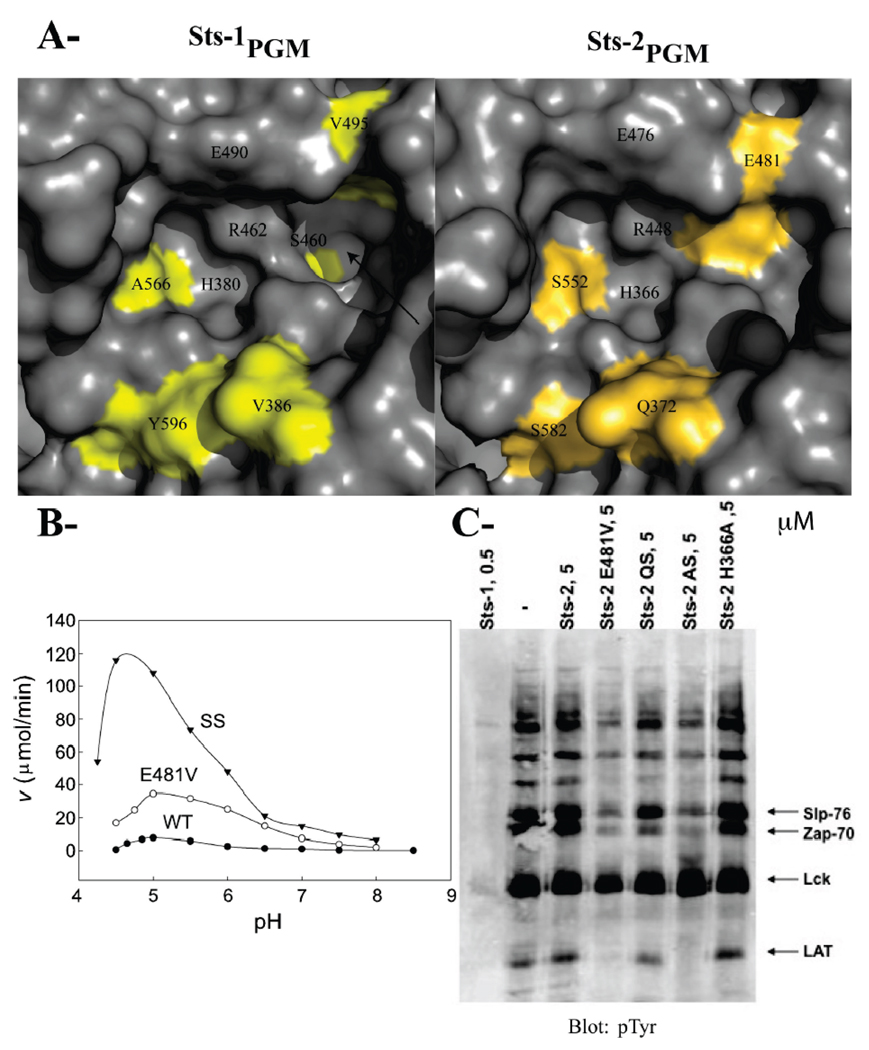
FIGURE 4. Residues that differentiate Sts-1PGM from Sts-2PGM.
(A) Surface representation of the active site of Sts-1PGM and Sts-2PGM. Nonconserved residues are colored yellow and orange, respectively. The hydrophobic cavity that is solvent-exposed in Sts-1PGM but blocked by Glu481 in Sts-2PGM is indicated with an arrow. (B) pH dependence of the phosphatase activity of Sts-2PGM. Initial velocities of pNPP (1 mM) hydrolysis by wild-type (●), E481V (○), and SS (S552A/S582Y) (▼) mutants of Sts-2PGM (1 µM) were measured and plotted at the indicated pH and 37 °C. (C) Proteins from TCR-stimulated Jurkat cells were isolated by immunoprecipitation, eluted from the pTyr antibodies, and evaluated as substrates for Sts-2PGM mutants at the indicated concentration in micromolar. QS is the Q372V/S582Y double mutant and AS the A446S/S552A double mutant. Arrows indicate the bands for the Slp-76, Zap-70, Lck, and LAT proteins. The assay was repeated at least three times, and the gel is representative of one experiment.