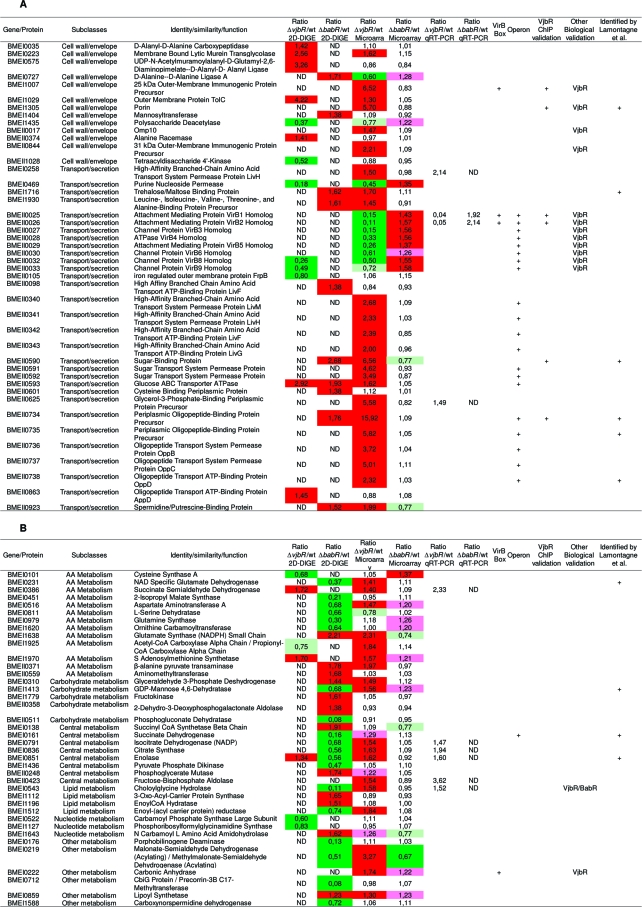
Table 2. Targets Identified in This Studya.
 |
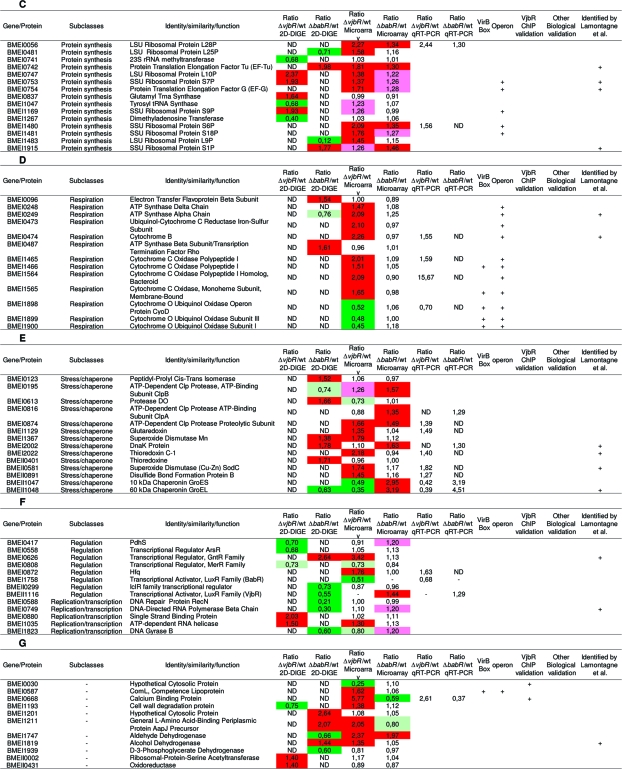 |
Summary table of targets genes identified in this study and connections with other published results. Each target is defined by a BMEnnnnn number (corresponding to the ORF number of the gene in Brucella melitensis 16 M genome), a functional class and a predicted function. A: Cell wall biogenesis and transport/secretion subclasses. B: Metabolism subclass. C: Translation subclass. D: Respiration process subclass. E: Stress response subclass. F: Regulation subclass. G: Unclassified targets. In the fold change column, colors represent the regulator’s effect: red when the regulator exerts a repressive role (fold change >1.3) and green when the regulator exerts an activation role (fold change <0.7). Light colors were used for genes with a lower fold change (pink: 1.3 > fold change >1.2; olive-green: 0.8 > fold change >0.7). Twenty-nine targets of interest were analyzed by qRT-PCR on new biological samples to validate microarray results. These results are listed in the “Ratio mutant/wt qRT-PCR” column. The “VirB Box” column indicates with a “+” genes containing in their promoter sequence the box identified by de Jong(35) for VjbR regulation. “Operon” column indicates genes which are predicted by BioCyc or KEGG DAS to be part of an operon. Positive results for VjbR ChIP experiments are labeled with a “+” in the “VjbR ChIP validation” column. When biological validations were available (such as Western blots, bile salts resistance test.. .), mutant strain’s name tested can been found in the “Biological validation” column. In the last column, genes identified by a “+” have been found by Lamontagne and coworkers(17) to be implicated in Brucella abortus intracellular adaptation. ND: not determined.
