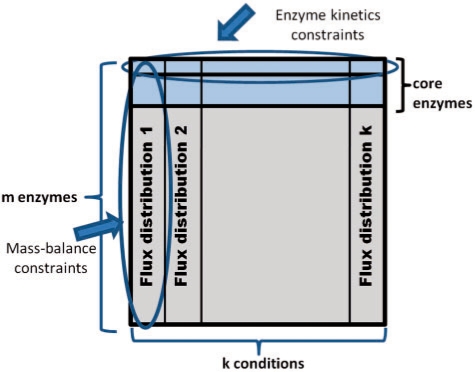
Fig. 1.
The figure illustrates the associations between variables in IOMA's optimization problem imposed by the various constraints. Rows represent enzymes and columns represent growth conditions—i.e. the j-th column representing the flux distribution for the j-th condition (denoted by 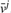 . Mass-balance [Equation (6)] and reaction directionality [Equation (7)] create dependencies between fluxes through different enzymes in one condition, irrespectively of all others conditions (i.e. associating fluxes in one column). The enzyme-kinetic constraint [Equation (8)] associates between fluxes through one enzyme in different growth conditions (via the enzyme's parameters vmax+ and vmax−, which are condition-invariant), irrespectively of all other enzymes (i.e. associating fluxes in one row). The latter constraint is defined only for a core set of enzymes for which metabolomic and proteomic data is available (marked in blue). The union of both types of row and column constraints in IOMA's optimization indirectly associates between many additional fluxes through various enzymes in different conditions.
. Mass-balance [Equation (6)] and reaction directionality [Equation (7)] create dependencies between fluxes through different enzymes in one condition, irrespectively of all others conditions (i.e. associating fluxes in one column). The enzyme-kinetic constraint [Equation (8)] associates between fluxes through one enzyme in different growth conditions (via the enzyme's parameters vmax+ and vmax−, which are condition-invariant), irrespectively of all other enzymes (i.e. associating fluxes in one row). The latter constraint is defined only for a core set of enzymes for which metabolomic and proteomic data is available (marked in blue). The union of both types of row and column constraints in IOMA's optimization indirectly associates between many additional fluxes through various enzymes in different conditions.