Abstract
Florida red tides occur as the result of blooms of the marine dinoflagellate Karenia brevis. K. brevis is known to produce several families of fused polyether ladder compounds. The most notable compounds are the brevetoxins, potent neurotoxins that activate mammalian sodium channels. Additional fused polyether ladder compounds produced by K. brevis include brevenal, brevisin, and hemi-brevetoxin B, all with varying affinities for the same binding site on voltage sensitive sodium channels. The structure elucidation and biological activity of two additional fused polyether ladder compounds containing seven fused ether rings will be described in this paper. Tamulamide A (MW=638.30) and tamulamide B (MW=624.29) were isolated from K. brevis cultures and their structures elucidated using a combination of NMR spectroscopy and high resolution mass spectrometry. Tamulamides A and B were both found to compete with tritiated brevetoxin-3 ([3H] PbTx-3) for its binding site on rat brain synaptosomes. However, unlike the brevetoxins tamulamide A and B showed no toxicity to fish at doses up to 200 nM and did not cause significant bronchoconstriction in sheep pulmonary assays.
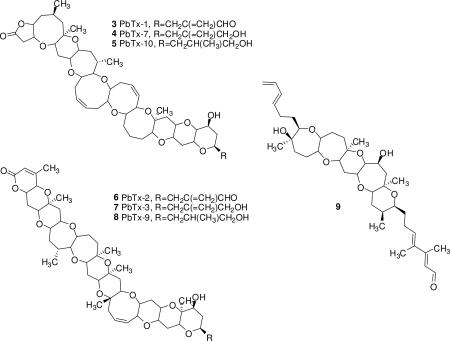
Karenia brevis is a marine dinoflagellate known for production of several different families of bioactive ladder frame polyether compounds; brevetoxin A backbone (3-5),1 brevetoxin B backbone (6-8),2 hemibrevetoxin B,3 brevenal (9),4,5 and brevisin.6 All of the above polyethers contain similar trans-fused, ladder-shaped, cyclic ether ring systems, however, ring sizes (5-9 membered rings), number of rings (4, 5, 6, 10 and 11) and side chains vary among the different families. The brevetoxin B family has the largest number of rings (11) and hemibrevetoxin B the fewest number of rings (4). Another similarity among most of the previously described natural polyethers isolated from K. brevis is that they are able to compete with tritiated brevetoxin 3 ([3H]-PbTx-3) for its binding site (site 5) on voltage sensitive sodium channels (VSSCs) in rat brain synaptosomes.4,6,7 Of the preceding polyethers, brevetoxin A and B families have the highest affinity (low nM range) to site 5 in VSSCs and brevisin has the lowest affinity (low μM range). The brevetoxins have been shown to be potent neurotoxins8-13 and brevenal has been found to be a functional antagonist to brevetoxin, inhibiting brevetoxin's activity in all assays tested.4,14 Not only has brevenal been shown to reduce brevetoxin binding to rat brain synaptosomes but it also blocks brevetoxin induced bronchoconstriction in sheep14 and attenuates brevetoxin toxicity in fish.4
In this report we describe the isolation, structural characterization, and biological activity of a new family of ladder-frame polyether compounds called tamulamides: tamulamide A (TamA) (1) and tamulamide B (TamB) (2). These new compounds contain a novel fused polyether backbone containing 7 fused cyclic ether rings (6,6,6,7,6,7,6-membered), an amide side chain similar to the cyclic alkaloid brevisamide15 and an aldehyde moiety similar but not identical to brevenal.4,5 Both compounds reduce [3H]-PbTx-3 binding at site 5 in rat brain synaptosomes, have low toxicity to fish and cause only slight bronchoconstriction in sheep. It is speculated that the tamulamides may have antagonistic properties at the brevetoxin binding site similar to brevenal.
TamA (1) was obtained as a colorless, non-crystalline compound. HRESI-TOFMS spectra showed the sodium adduct [M + Na]+ at m/z 662.2934 which corresponded to the molecular formula of C35H45NO10Na and 14 double bond equivalents. The 1H NMR spectrum revealed the presence of 45 protons and an aldehyde group, similar to but different from that of brevenal,4 due to the presence of a doublet with a chemical shift of 9.55 ppm instead of a doublet with a chemical shift of 10.18 ppm as seen in brevenal. HSQC, DEPT, and 13C NMR experiments were able to verify the existence of 35 carbons. 1H and 13C NMR assignments are listed in Table 1.
Table 1.
NMR Spectroscopic Data (500 MHz, CD2Cl2) for TamA and TamB.
| position | TamA δC, mult. | δH mult. (J in Hz). | TamB δC, mult. | δH mult. (J in Hz). |
|---|---|---|---|---|
| 1 | 23.1, CH3 | 2.01, s | 23.1, CH3 | 2.00, s |
| 2 | 172.7, qC | -- | 172.7, qC | -- |
| 3 | 40.6, CH2 | 3.11, m | 40.6, CH2 | 3.09, m |
| 3.92, m | 3.93, m | |||
| 4 | 82.6, CH | 3.12, m | 82.4, CH | 3.08, m |
| 5 | 65.6, CH | 3.31, m | 65.4, CH | 3.30, m |
| 6 | 37.1, CH2 | 1.51, t (12) | 36.9, CH2 | 1.51, t (11) |
| 2.30, dt (11, 4) | 2.30, m | |||
| 7 | 79.2, CH | 3.06, m | 79.0, CH | 3.03, dt (17, 2) |
| 8 | 77.5, CH | 3.22, dd (13, 4) | 77.2, CH | 3.20. m |
| 9 | 43.4, CH2 | 1.45, t (11) | 43.5, CH2 | 1.42, t (12) |
| 2.07, dd (11, 4) | 2.11, t (11) | |||
| 10 | 73.6, qC | -- | 72.9qC | -- |
| 11 | 79.1, CH | 3.23, m | 78.8, CH | 3.09, m |
| 12 | 39.4, CH2 | 1.69, t (13) | 32.5, CH2 | 1.62, t (12) |
| 1.79, dd (12, 4) | 2.05, dd (11,5) | |||
| 13 | 75.7, qC | -- | 79.6, CH | 3.33, m |
| 14 | 75.3, CH | 4.38, br s | 73.1, CH | 4.17, d (9) |
| 15 | 131.5, CH | 5.45, dt (12, 3) | 133.0, CH | 5.61, dt (8, 4) |
| 16 | 131.6, CH | 5.64, dt, (12, 2) | 131.5, CH | 5.67, m |
| 17 | 80.4, CH | 3.84, m | 80.4, CH | 3.83, m |
| 18 | 67.4, CH | 3.63, m | 77.56, CH | 3.34, m |
| 19 | 38.9, CH2 | 1.53, t (12) | 38.8, CH2 | 1.54, t (12) |
| 2.15, q, (4) | 2.29, m | |||
| 20 | 77.7, CH | 3.32, m | 77.78, CH | 3.28, m |
| 21 | 80.4, CH | 3.82, m | 80.3, CH | 3.81, m |
| 22 | 132.5, CH | 5.77,dt (13, 3) | 132.5, CH | 5.77, dt (12, 3) |
| 23 | 132.1, CH | 5.67, dt (13, 2) | 131.9, CH | 5.68, m |
| 24 | 77.6, CH | 4.10, dt (5, 3) | 77.68, CH | 4.09, m |
| 25 | 76.2, CH | 3.87, m | 76.1, CH | 3.87, m |
| 26 | 128.76, CH | 5.73, t (8) | 128.71, CH | 5.72, t (8) |
| 27 | 128.85, CH | 5.75, t (8) | 128.74, CH | 5.74, t (8) |
| 28 | 74.74, CH | 4.80, br s | 74.6, CH | 4.79, br s |
| 29 | 142.3, CH | 6.16, q (8) | 142.1, CH | 6.16, q (6) |
| 30 | 128.9, CH | 6.57, dd (16, 11) | 128.6, CH | 6.56, dd (16, 12) |
| 31 | 151.3, CH | 7.11, dd (15, 11) | 151.0, CH | 7.10, dd (15, 11) |
| 32 | 132.6, CH | 6.14, q (8) | 132.4, CH | 6.13, q (9) |
| 33 | 193.8, CH | 9.55, d (8) | 193.9, CH | 9.54, d (8) |
| 34 | 15.6, CH3 | 1.26, s | 15.7, CH3 | 1.25, s |
| 35 | 17.0, CH3 | 1.27, s |
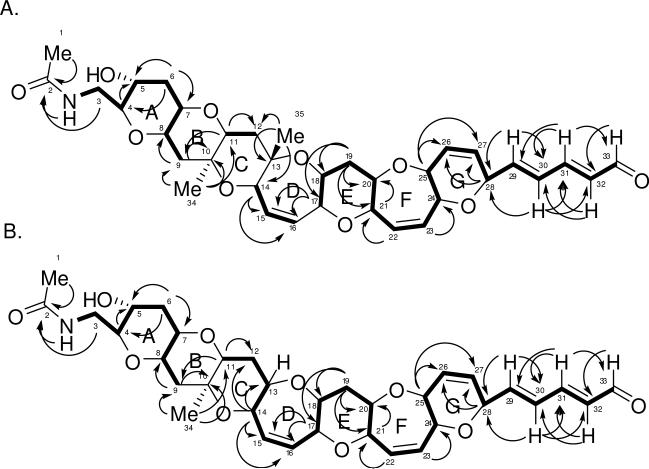
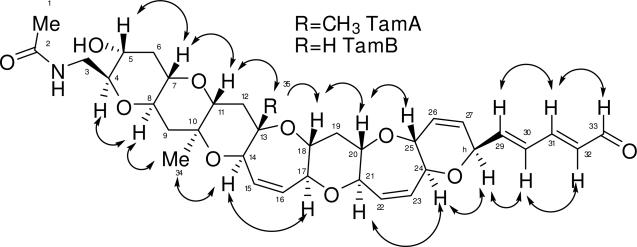
TOCSY NMR data was used to verify the various spin systems as seen in Figure 1A. The existence of the six-member ring with the double bond (ring G) was initially unclear due to the fact that protons on C-25 (3.87 ppm) and C-28 (4.80 ppm) had strong long-range coupling correlations in the COSY NMR experiment. However, the carbon and proton chemical shifts (see Table 1) indicate that both protons must be connected to a carbon bearing an oxygen atom. The relative stereochemistry of TamA was identified using ROESY correlations (Figure 2). The ROSEY correlations also helped to confirm the orientation of the ring G which was unclear due to extensive long range COSY correlations of protons C26 and C27, and also indicated the trans-syn orientation of the protons on the rest of the polyether backbone. Based on the structures of other polyether ladder compounds such as; brevetoxin,1,2 brevenal,4,5 ciguatoxin,16 gambierol,17 and gambieric acids,18 we anticipated that the vicinal oxymethine protons would have a trans relative orientation. The positions of the ether rings were deduced using the ether methines along the linear skeleton. Earlier work with polyethers18 confirmed ether bonds using ROE correlations between ether methines (angular protons). If the structure proposed for TamA is correct, there should be ROESY correlations between the protons on C-4 and C-8 (across ring A), the protons on C-7 and C-11 (across ring B), and so forth along the linear skeleton.
Figure 1.
TOCSY and HMBC correlations for A. TamA and B. TamB. Bold lines indicate spin systems as determined by 1H-1H correlations for TOCSY experiments. Arrows indicate 1H-13C correlations for HMBC experiments.
Figure 2.
1H-1H ROESY correlations for TamA and TamB. Double headed arrows indicate important ROESY correlations for TamA and TamB as determined by NMR. Single headed arrow indicates an additional ROESY correlation for TamB.
The ROESY NMR data did in fact prove these correlations. Confirmation of the orientation of the G-ring was obtained through ROESY correlations between the proton on C-28 (4.80 ppm) and the proton on C-24 (4.10 ppm) across ring G and the correlations between the proton on C-25 (3.87 ppm) and the proton on C-20 (3.32 ppm) across ring F. Some correlations were not seen due to overlapping signals in the proton spectrum (the protons on C-17 and C-21, for example).
HMBC correlations for TamA are shown in Figure 1A. The HMBC NMR data was especially useful in determining the placement of the three methyl groups and the placement of the three quaternary carbons (C-2, C-10, and C-13). Protons from the first methyl (protons on carbon 1, 2.01 ppm) were correlated to a carbon with a chemical shift of 172.7 ppm, confirming the presence of an amide carbonyl. The proton on the nitrogen (5.93 ppm) and the protons from C-3 (3.11 ppm, 3.92 ppm) are also correlated to C-2, confirming the amide containing side chain (C-1 to C-3). The second methyl (protons on C-34, 1.26 ppm) had correlations to C-9, C-10 (quaternary), and C-11. The third methyl (protons on C-35, 1.27 ppm) had correlations to C-12, C-13 (quaternary), and C-14. HMBC correlations also helped to confirm the placement of the carbons on ring G, specifically the protons on C-25 and C-28 correlated to C-26 and C-27. Other important correlations confirmed the placement of the two remaining sets of protons (on C-15 and C-16 and on C-22 and C-23) with overlapping signals resonating between 5 and 6 ppm that are attached to alkenes. The protons on C-14 and C-17 correlated to the alkene C-15 and C-16 (ring D) while the proton on C-22 was correlated to C-21 and the proton on C-23 was correlated to C-24 (ring F). The HMBC and TOCSY NMR data also revealed correlations demonstrating the presence of the double bond-containing chain extending from ring G to the aldehyde group (C-28 to C-33).
TamB (2) was obtained as a colorless, non-crystalline compound. HRESI-TOFMS spectra showed the sodium adduct [M + Na]+ at m/z 448.2781 which corresponded to the molecular formula of C34H43NO10Na and 14 double bond equivalents. The 1H NMR spectrum revealed the presence of 43 protons and an aldehyde group similar to that of TamA with a doublet at 9.55 ppm. HSQC, DEPT, and 13C NMR data were able to verify the existence of 34 carbons. Proton and carbon NMR assignments are listed in Table 1.
The proton, COSY, TOCSY, and HSQC NMR data of TamB revealed a structure very similar to TamA and HRMS revealed a difference of 14 amu in the molecular weight. The TOCSY NMR experiments were able to verify the spin systems as seen in Figure 2. The structure of TamB was found to be identical to TamA from C-1 to C-9 and C-19 to C-33.
The 13C, DEPT, HMBC, and HSQC NMR data of TamB revealed one fewer carbon than TamA, specifically one carbon missing within 0 and 25 ppm, and also revealed the existence of only two quaternary carbons (C-2 and C-10). This helped to confirm that a single proton with a chemical shift of 3.33 ppm replaced the methyl group that was attached to C-13 in TamA. HMBC NMR correlations between the proton on C-14 (4.17 ppm) and C-13 (79.6 ppm, not a quaternary carbon) along with correlations between the proton on C-14 and C-12 (32.5 ppm) add additional confirmation (Figure 1B). Carbons C-11 and C-18 and their respective protons were shifted slightly but proton-proton correlations were seen in the COSY, TOCSY (Figure 1B), and HSQC NMR data (see Table 1).
ROESY NMR data for TamB again demonstrated the position of the ether rings by correlating the ether methines along the linear skeleton. The relative stereochemistry identified using 1H-1H ROESY correlations can be seen in Figure 2. An important correlation was between the proton on C-11 (3.09 ppm) and the proton on C-13 (3.33 ppm) across ring C (single headed arrow), helping to prove that the methyl group from compound TamA was replaced by a proton with a chemical shift of 3.33 ppm in compound TamB. Some correlations were not seen due to overlapping signals in the proton spectrum (the protons on C-13 and C-18 and the protons on C-17 and C-21, for example). The ROESY data also revealed that TamB possesses a trans-syn fused ring system on its polycyclic backbone, with oxygen atoms alternating between the top and bottom, which is similar to the brevetoxins.1,2,19
Fish bioassay
Tamulamide A: There were no deaths in the fish that were exposed to 200, 100, or 50 nM TamA, however, there was one fish exposed to 25 nM TamA that died overnight (20 hours and 40.27 minutes). No vehicle controls (EtOH only) died and all positive controls (200 nM PbTx-2) died within 29.50 ± 5.18 minutes (n=5). Administration of TamB resulted in no deaths for the fish at any dose. No vehicle controls (EtOH only) died, but all positive controls (200 nM PbTx-2) died within 46.72 ± 4.61 minutes (n=5).
Sheep bioassay
The bronchoconstrictor responses seen after airway challenge with TamA and TamB are shown in Table 2. Administration of TamA increased lung resistance (rL) by 40 ± 2% while the average bronchoconstrictor response of TamB was an increase in rL of 35 ± 1%, as compared to saline controls. For comparison, a previous study by Abraham et al.14 determined that airway challenge with 20 breaths of a 10 pg/mL treatment of PbTx-2 produced an increase in rL of 226 ± 21% and a 10 pg/mL treatment of PbTx-3 produced an increase in rL of 179 ± 22%. Alternatively, the functional antagonist brevenal produced no constrictor response.
Table 2.
Effects of TamA and TamB on Pulmonary airflow resistance (rL) in sheep.
| TamA - 10 pg/ml | |||
|---|---|---|---|
| Dose : 20 breaths given aerosol | |||
| | |||
| Sheep# | Baseline | TamA | |
| | |||
| rL | rL | % | |
| 1988 | 0.99 | 1.38 | 39% |
| 2164 | 0.93 | 1.33 | 43% |
| 2263 | 0.96 | 1.36 | 42% |
| 2297 | 1.00 | 1.34 | 34% |
| Means: | 0.97 | 1.35 | 40% |
| S.D. : | 0.03 | 0.02 | 4% |
| S.E. : | 0.02 | 0.01 | 2% |
| TamB - 10 pg/ml | |||
|---|---|---|---|
| Dose : 20 breaths given aerosol | |||
| | |||
| Sheep# | Baseline | TamB | |
| | |||
| rL | rL | % | |
| 1988 | 1.00 | 1.36 | 36% |
| 2164 | 0.94 | 1.26 | 34% |
| 2263 | 0.97 | 1.33 | 37% |
| 2297 | 0.99 | 1.32 | 33% |
| Means: | 0.98 | 1.32 | 35% |
| S.D. : | 0.03 | 0.04 | 2% |
| S.E. : | 0.01 | 0.02 | 1% |
Receptor binding assay
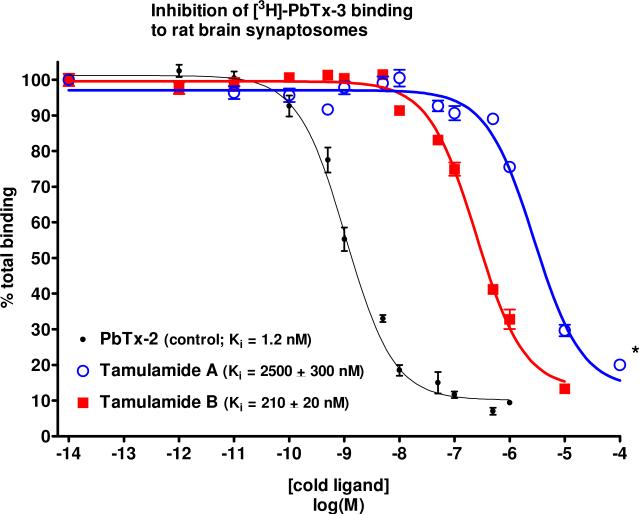
The results of the receptor binding assay with TamA and TamB can be seen in Figure 3. TamB was able to displace tritiated brevetoxin ([3H]-PbTx-3) at high nanomolar concentrations with an apparent Ki (Ki is the concentration of a ligand needed to displace one half of the radio-ligand from its binding site) of 210 ± 20 nM while TamA displaced [3H]-PbTx-3 at low micromolar concentrations, apparent Ki = 2.5 ± 0.3 μM. The concentration required for TamB to compete for sites occupied by brevetoxin was two orders of magnitude greater than for PbTx-2, while the concentration required for TamA to compete for sites occupied by brevetoxin was over three orders of magnitude greater than for PbTx-2. ANOVA revealed that the Ki for TamB was significantly different from the Ki for TamA (P<0.001) and the Ki values for both TamA and TamB were significantly different from the Ki value for the PbTx-2 control (1.2 nM, P<0.001).
Figure 3.
Inhibition of [3H]-PbTx-3 binding to rat brain synaptosomes by TamA and TamB. Each data point represents the mean ± S.E.M. of at least 3 experiments. * indicates two experiments at the indicated concentration due to lack of material.
Discussion
Two novel compounds TamA and TamB, each with a seven polyether ring system containing an aldehyde side chain, have been isolated and identified from cultures of K. brevis. In addition to establishing the structures of the two novel compounds isolated from K. brevis, the biological activity of the two novel compounds was determined using both in vivo and in vitro bioassays.
In vivo assays suggest low toxicity for TamA and TamB. Administration of TamA and TamB to fish revealed that these compounds had low 24 hr toxicity in the dose range (0-200 nM) at which they were administered. TamA and TamB were than tested for their bronchoconstrictor effects in a sheep model of asthma. Airway challenge with TamA and TamB resulted in slight increases in rL (35-40%). The changes seen with TamA and TamB were 75-85 % less severe when compared with the changes seen with equivalent doses (200 pg) of PbTx-2 and PbTx-3.14 Although minor, the responses to TamA and TamB were significantly higher than those previously found for the brevetoxin antagonists, brevenal and β-naphthoyl-brevetoxin, which produced increases in rL of 4 ± 2% and 0 ± 3%, respectively.14 Therefore, compared to brevetoxins, the increases produced by TamA and TamB were attenuated (<40% for TamA and TamB as compared to >150% for the brevetoxins) but not without activity as seen with the brevetoxin antagonists brevenal and β-naphthoyl-brevetoxin. The results of this assay suggest that TamA and TamB, although partial agonists in this assay system, are most likely not major factors in the bronchoconstriction observed in humans inhaling Florida red tide aerosols. Future studies will determine if either TamA or TamB have any inhibitory effects on the bronchoconstrictor response of brevetoxins, as reports on brevenal and β-naphthoyl-brevetoxin have shown.
The results of the receptor binding assays with TamA and TamB are noteworthy, considering the tenfold loss in binding affinity with the extra methyl group in TamA. From our results it appears that the presence of the methyl group interferes with binding. A hypothesis for the difference in binding between the two compounds is that the extra methyl group in TamA may lock the overall structure of the molecule into a specific confirmation, not allowing the molecule to fit correctly into the binding region. A second hypothesis is that the extra methyl group provides steric hindrance that is physically blocking the molecule from fitting correctly into the binding region. Preliminary molecular modeling provides evidence for the second proposal (data not shown).
Also of note is the fact that TamB displaced brevetoxin from its binding site on the same order of magnitude as brevenal (Ki = 170 nM). Because brevenal reduces tritiated brevetoxin binding to site 5 of VSSCs and it has been shown to be non-toxic and to protect fish from the effects of brevetoxin in fish bioassays, it has been deemed a natural functional antagonist of brevetoxin.4 Given that TamB reduces tritiated brevetoxin binding to site 5 of VSSCs at similar concentrations as brevenal and has been shown to be non-toxic in various bioassays, this novel compound has the potential to act as a second natural functional antagonist to brevetoxin.
Experimental Section
General Experimental Procedures
Optical rotation was measured with a Rudolph Research Analytical Autopol III automatic polarimeter. The maximum UV absorption was measured using a Molecular Devices Flexstation III multimode plate reader. NMR spectra were acquired on a Bruker Avance 500 MHz spectrometer and referenced to residual solvent 1H and 13C signals (δH 5.32, δC 54.00 for CDCl2) using a TXI 1.7 mm probe. An ABI Sciex QSTAR XL MS/MS QTOF mass spectrometer in positive ionization mode was used to determine the accurate (±0.3 ppm) molecular weight, the chemical formula, and the number of double bond equivalents. Purification of the compounds was carried out using a series of liquid-liquid and solid phase separation techniques. Liquid-liquid separation was achieved using a Kromaton 200 fast centrifugal partitioning chromatography system (FCPC) attached to an ISCO 2350 solvent delivery pump. Fractions from the Kromaton were spotted onto 5 cm × 6.5 cm aluminum foil silica gel (60 F254) to determine which fractions the target compounds were in. Final separation and purification of compounds was accomplished using high performance liquid chromatography (HPLC) using a Shimadzu LC-10A system controller, SPD-10AV UV-VIS dual wavelength detector (215 and 295 nm) and 2 LC-10AS HPLC pumps. Purification of the compounds was carried out using two different HPLC columns. Initially, a Varian Dynamax microsorb 100Å, 5 μm, 10 × 250 mm column was used followed by a Phenomenex Luna phenyl-hexyl, 100Å, 5 μm, 4 × 250 mm column for final purification.
Isolation of TamA and TamB
Cultures of K. brevis (100 L, Wilson's 58 clone) were extracted with CHCl3 after 30 days of growth. The CHCl3 layer was collected and dried under vacuum. The CHCl3 fraction was then partitioned between H2O and EtOAc (50:50, v:v). The EtOAc layer was collected and dried under vacuum, and then partitioned between petroleum ether:MeOH:H2O (50:40:10, v:v:v). The MeOH/H2O layer was collected and dried under vacuum, and an initial separation with an FCPC. was achieved using a three step gradient.. Prior to injection of the extract onto the FCPC, the stationary phase (MeOH and H2O, (2:1, v:v)) was run through the Kromaton for 30 minutes at 500 RPM (10 mL/min). A solution of the stationary phase was used to saturate the mobile phase for each step of the gradient by adding equal volumes of the mobile and stationary phases to a flask and sonicating the mixture of solutions for 1 minute. After sonication the layers were allowed to separate and the stationary phase was removed, the remaining mobile phase was then introduced to the Kromaton for the gradient steps. Gradient step 1: The mobile phase was hexane and EtOAc (2:0.1, v:v). Gradient step 2: The mobile phase was hexane and EtOAc (1:1, v:v). Gradient step 3: The mobile phase was hexane and EtOAc (2:1, v:v). Prior to injection onto the FCPC the sample was dissolved in 3 mL of MeOH and H2O (2:1, v:v) and 3 mL of hexane and EtOAc (2:0.1, v/v), and then injected into the Kromaton. The instrument was run at 1200 rpm at 5 mL/min for 30 minutes in each gradient step. After completion of the gradient series the Kromaton was then flushed for another 50 minutes at 250 rpm at 5 mL/min using methanol and water (2:1, v:v). A total of 72 fractions were collected in 18 × 150 mm glass test tubes. Three minute fractions were collected for gradient steps 1, 2, and the flush. Two minute fractions were collected for gradient step 3. Samples of each fraction were applied to TLC plates and eluted with CH2Cl2 and MeOH (95:5, v:v). Bands were visualized using H2SO4/H20 (25:75 v:v) and similar fractions combined. TamA and TamB were found in the most polar fractions (61-72) eluting from the Kromaton.
The Kromaton fractions containing TamA and TamB were combined, filtered using a nylon 0.22 μm disc filter, and then dried under vacuum. Separation of TamA and TamB from other compounds in the Kromaton fractions was achieved using semi-preparative reversed phase HPLC (Dynamax column) with a binary solvent system (MPA-100% H20, MPB-100% MeOH). The elution gradient was as follows: 30% MPA to 0% MPA over 20 min at a flow rate of 3.4 mL/min. TamA (tR=13.5min) and TamB (tR=13.0min) were collected separately and further purified using reversed-phase HPLC (Phenomenex column) (MeOH/H20, 85/15, 1.4 mL/min) this process yielded 0.7 mg TamA (tR=6.5min) and 0.5mg TamB (tR=6.0min) from 100 L of culture with a purity greater that 98% as measured by NMR and HPLC-UV.
Tamulamide A (1): colorless, non-crystalline; [α]22D=95.5 (c 0.1, ACN); UV (ACN) λmax (log ε) 286 (3.30); for 1H and 13C NMR data see table 1; HRESI-TOFMS m/z 662.2934 ([M + Na]+ (calcd for C35H45NO10Na 662.2941).
Tamulamide B (2): colorless, non-crystalline; [α]22D=105.5 (c 0.1, ACN); UV (ACN) λmax (log ε) 290 (3.11); for 1H and 13C NMR data see table 1; HRESI-TOFMS m/z 648.2781 ([M + Na]+ (calcd for C34H43NO10Na 648.2785).
Fish Bioassay
Mosquito fish (Gambusia affinis) were placed in 50-mL beakers containing 20 mL of fish tank water. One hundred μg of each test compound (TamA, TamB and PbTx-2 (positive control)) were dissolved in 1 mL of EtOH and then TamA and TamB were serially diluted in EtOH to the following stock concentrations (500 nM, 1000 nM, 2000 nM, 4000 nM). Fish were then administered 0.2 mL of one of the test solutions above or vehicle control (0.2 mL EtOH) that resulted in final concentrations of 0 nM, 25 nM, 50 nM, 100 nM, or 200 nM in the water. The PbTx-2 control concentration was 200 nM. Five fish were used at each concentration (30 fish total). After addition of the compounds, the fish were monitored for 24 hours or until time of death. Time of death (in minutes) is given as mean ± S.E.M. Significant differences were determined using a two-way Student's t-test.
Sheep Bioassay
The methods for determining pulmonary airflow resistance (rL) values have been described in detail.20 Briefly, a balloon catheter was advanced through one nostril into the lower esophagus and the animals were intubated with a cuffed endotracheal tube through the other nostril. Animals remained intubated throughout the course of a particular experiment, but to avoid discomfort during these studies, the cuff of the endotracheal tube was inflated only during the measurements of pulmonary resistance (rL) and during delivery of nebulized agents. Pleural pressure was measured via the esophageal catheter. Lateral pressure in the trachea was measured with a sidehole catheter advanced through and positioned distal to the tip of the endotracheal tube. Transpulmonary pressure, the difference between tracheal and pleural pressure, was measured with a differential pressure transducer. To measure pulmonary resistance (RL), the proximal end of the endotracheal tube was connected to a pneumotachograph and the signals of flow and transpulmonary pressure were recorded on a computer. Respiratory volume was obtained by digital integration of the flow signal so that RL was calculated from the transpulmonary pressure and flow, at isovolumetric points. Analysis of 5-10 breaths was used for each determination of RL. The rL values were determined before (baseline) and immediately after airway challenge with TamA and TamB (n=4) and a percent change from baseline was calculated. The sheep were challenged with 20 breaths of a 10 pg/mL concentration of each compound. The rL values for each compound are presented as mean ± S.E.M. Significant differences were determined using a two-tailed T-test.
Receptor Binding Assay
Competitive rat brain synaptosome binding assays were performed using a rapid filtration technique.7,21 All binding experiments were performed on ice using 0.5 mL total volume of Standard Binding Medium (SBM; 50 mM HEPES, 130 mM choline chloride, 5.4 mM KCl, 0.8 mM MgSO4-7H2O, 5.5 mM glucose, 1 mM EGTA, 0.01% Emulphor EL-620 emulsifier; pH 7.4) containing 1 mg/mL bovine serum albumin (SBM+BSA). Competition for binding to rat brain synaptosomes versus [3H]-PbTx-3 (approximately 1.5 nM) was performed using PbTx-2 (control), TamA, and TamB (competitor concentrations ranged from 0 to 10 μM). Synaptosome mixture (0.1 mg protein/mL final concentration), cold ligand, [3H]-PbTx-3, and binding medium were allowed to incubate on ice in 96-well assay plates for at least one hour. Synaptosomes were separated from the other components by rapid filtration onto 96-well filter plates (Packard Unifilter GB, 1 μM filter membrane) using a Packard Filtermate Cell Harvester. After addition of scintillation cocktail (Packard Microscint 20), the amount of [3H]-PbTx-3 retained by the synaptosomes was determined by liquid scintillation spectrometry (Packard TopCount NXT). Nonlinear regression analysis (one-site competition) was performed using GraphPad Prism version 4.03 for Windows, GraphPad Software (San Diego, CA), www.graphpad.com. All derived values were the mean of at least 3 experiments (except where indicated), with duplicate or triplicate determination at each concentration of inhibitor in each experiment. An ANOVA followed by a Newman-Keuls multiple comparison post test was used for statistical analysis of apparent equilibrium inhibition (Ki) values.
Supplementary Material
Acknowledgement
This work was supported by grant P01 ES10594 from NIEHS. Thanks to S. Niven, S. Carvalho, and T. Hogue for maintaining cultures of Karenia brevis. Special thanks to Dr. J.L.C. Wright and Dr. R. Van Wagoner for help with structure elucidation and to Dr. Van Wagoner for running mass spectrometry.
Footnotes
Supporting Information Available: NMR spectra for tamA (1) and tamB (2). This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Shimizu Y, Chou H-Y, Bando H. J. Am. Chem. Soc. 1986;1089:514–515. doi: 10.1021/ja00263a031. [DOI] [PubMed] [Google Scholar]
- 2.Lin Y, Risk M, Ray SM, Van Engen D, Clardy J, Golik J, James JC, Nakanishi KJ. J. Am. Chem. Soc. 1981;103:6773–6775. [Google Scholar]
- 3.Prasad AVK, Shimizu Y. J. Am. Chem. Soc. 1989;111:6476–6477. [Google Scholar]
- 4.Bourdelais AJ, Jacocks H, Wright JLC, Bigwarf PM, Baden DG. J. Nat. Prod. 2005;68:2–6. doi: 10.1021/np049797o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuwa H, Ebine M, Bourdelais AJ, Baden DG, Sasaki M. J. Am. Chem. Soc. 2006;128:16989–16999. doi: 10.1021/ja066772y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satake M, Campbell A, Van Wagoner RM, Bourdelais AJ, Jacocks H, Baden DG, Wright JLC. J. Org. Chem. 2009;74:989–994. doi: 10.1021/jo802183n. [DOI] [PubMed] [Google Scholar]
- 7.Poli MA, Mende TJ, Baden DG. Mol. Pharmacol. 1986;30:129–135. [PubMed] [Google Scholar]
- 8.Baden DG, Mende TJ. Toxicon. 1982;20:457–461. doi: 10.1016/0041-0101(82)90009-5. [DOI] [PubMed] [Google Scholar]
- 9.Huang JMC, Wu CH, Baden DG. J. Pharmacol. Exp. Ther. 1984;229:615–627. [PubMed] [Google Scholar]
- 10.Ishida Y, Shibata S. Pharmacology. 1985;31:237–240. doi: 10.1159/000138120. [DOI] [PubMed] [Google Scholar]
- 11.Borison HL, McCarthy LE, Ellis S. Toxicon. 1985;23:517–524. doi: 10.1016/0041-0101(85)90036-4. [DOI] [PubMed] [Google Scholar]
- 12.Dickey R, Jester E, Granade R, Mowdy D, Moncrief C, Rebarch KD, Robl M, Musser S. Poli, M. Nat. Tox. 2000;7:157–165. doi: 10.1002/(sici)1522-7189(199907/08)7:4<157::aid-nt52>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 13.Poli MA, Musser SM, Dickey RW, Eilers PP, Hall S. Toxicon. 2000;38:981–993. doi: 10.1016/s0041-0101(99)00191-9. [DOI] [PubMed] [Google Scholar]
- 14.Abraham WM, Bourdelais AJ, Sabater JR, Ahmed A, Lee TA, Serebriakov I. Am. J. Respir.y and Crit. Care Med. 2005;171:26–34. doi: 10.1164/rccm.200406-735OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satake M, Bourdelais AJ, Van Wagoner RM, Baden DG, Wright JL. Org. Lett. 2008;10:3465–3468. doi: 10.1021/ol801243n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murata M, Legrand AM, Ishibashi Y, Fukui M, Yasumoto T. J. Am. Chem. Soc. 1990;112:4380–4386. [Google Scholar]
- 17.Satake M, Murata M, Yasumoto T. J. Am. Chem. Soc. 1993;115:361–362. [Google Scholar]
- 18.Nagai H, Murata M, Torigoe K, Satake M, Yasumoto T. J. Org. Chem. 1992;57:5448–5453. [Google Scholar]
- 19.Gawley RE, Rein KS, Kinoshita M, Baden DG. Toxicon. 1992;30:780–785. doi: 10.1016/0041-0101(92)90014-v. [DOI] [PubMed] [Google Scholar]
- 20.Abraham WM, Sielczak MW, Ahmed A, Cortes A, Lauredo IT, Kim J, Pepinsky B, Benjamin CD, Leone DR, Lobb RR. J. Clin. Invest. 1994;93:776–787. doi: 10.1172/JCI117032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dodd PR, Hardy JA, Oakley AE, Edwardson JA, Perry EK, Delaunoy JP. Brain Res. 1981;226:107–118. doi: 10.1016/0006-8993(81)91086-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.