Abstract
Aminomethyltransferase, a component of the glycine cleavage system termed T-protein, reversibly catalyzes the degradation of the aminomethyl moiety of glycine attached to the lipoate cofactor of H-protein, resulting in the production of ammonia, 5,10-methylenetetrahydrofolate, and dihydrolipoate-bearing H-protein in the presence of tetrahydrofolate. Several mutations in the human T-protein gene are known to cause nonketotic hyperglycinemia. Here, we report the crystal structure of Escherichia coli T-protein in complex with dihydrolipoate-bearing H-protein and 5-methyltetrahydrofolate, a complex mimicking the ternary complex in the reverse reaction. The structure of the complex shows a highly interacting intermolecular interface limited to a small area and the protein-bound dihydrolipoyllysine arm inserted into the active site cavity of the T-protein. Invariant Arg292 of the T-protein is essential for complex assembly. The structure also provides novel insights in understanding the disease-causing mutations, in addition to the disease-related impairment in the cofactor-enzyme interactions reported previously. Furthermore, structural and mutational analyses suggest that the reversible transfer of the methylene group between the lipoate and tetrahydrofolate should proceed through the electron relay-assisted iminium intermediate formation.
Keywords: Enzyme Catalysis, Enzyme Structure, Folate, Protein Assembly, X-ray Crystallography, Aminomethyltransferase, Glycine Cleavage System, H-protein, Nonketotic Hyperglycinemia
Introduction
The glycine cleavage system (GCS)4, a multienzyme system composed of four proteins termed P-, H-, T-, and L-protein, catalyzes the reversible oxidation of glycine yielding carbon dioxide, ammonia, 5,10-methylenetetrahydrofolate (5,10-CH2-THF), and a reduced pyridine nucleotide (1). The functional importance of this system is underscored by the fact that GCS deficiencies are linked to nonketotic hyperglycinemia (NKH), a metabolic disorder with autosomal recessive inheritance characterized by a massive accumulation of glycine in body fluids, causing severe, frequently lethal, neurological symptoms in the neonatal period. The enzymatic analysis of NKH patients revealed that ∼85% have a P-protein deficiency, and the rest have a T-protein deficiency (2). We have previously identified three mutations in human T-protein (huT) that caused NKH (3), and a total of 12 mutations have been reported in the T-protein gene of patients in the Human Gene Mutation Data Base, Cardiff, UK. Our crystal structure of huT revealed that five of nine disease-related missense mutations are localized around the folate cofactor, forming extensive hydrogen bond networks supporting the cofactor (4). On the other hand, some mutations are localized near the N terminus of the protein, which has been speculated to be involved in the protein-protein interaction with H-protein (5, 6).
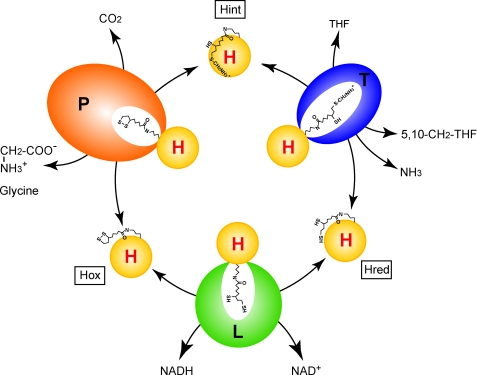
The overall reaction catalyzed by GCS proceeds in three steps (Fig. 1). First, P-protein (glycine dehydrogenase; EC 1.4.4.2) decarboxylates glycine in a pyridoxal phosphate-dependent reaction, and the remaining aminomethyl moiety is passed onto the distal sulfur (S8) atom of the lipoate cofactor of H-protein. Second, T-protein (aminomethyltransferase; EC 2.1.2.10) degrades the aminomethyl moiety to ammonia and 5,10-CH2-THF in the presence of THF, leaving dihydrolipoate-bearing H-protein (Hred). Finally, L-protein (dihydrolipoamide dehydrogenase; EC 1.8.1.4) catalyzes the reoxidation of the reduced lipoate cofactor in the presence of pyridine nucleotide. A unique feature of GCS is the fact that the H-protein, having a covalently attached R-(+)-lipoic acid, is engaged as a mobile substrate that commutes successively between the three enzymes. GCS also catalyzes the synthesis of glycine from 5,10-CH2-THF, ammonia and carbon dioxide in a reverse reaction (7). In the absence of THF, formaldehyde is generated instead of 5,10-CH2-THF (8). Although three-dimensional structures of individual components from various sources have already been reported (4, 9–16), no information concerning the complex structures of these components is available, except our cross-linking study (5), the structure of Thermotoga maritima T-protein (tmT) complexed with a protein-free dihydrolipoic acid (11), and a two-dimensional NMR study of pea H-protein (peaH) showing chemical shift changes upon T-protein binding (17). The crystal structure of peaH showed that the lipoyllysine arm gets docked into the binding cleft of the H-protein after aminomethyl loading (9). On the other hand, T-protein has a cloverleaf-like structure consisting of three domains with a central folate-binding cavity (4, 10, 11). The folate cofactor enters into the funnel-shaped cavity from the C-terminal wider opening and binds to the cavity in a kinked shape with the N5 and N10 atoms pointing to the N-terminal side at ∼13–14 Å in depth from the surface (see supplemental Fig. S6A). These structures raise the question how the aminomethyl lipoate arm leaves the binding cleft on H-protein and reaches the folate cofactor through the narrower opening of T-protein cavity.
FIGURE 1.
Outline of the reversible reaction of the GCS. P, T, L, and H are the protein components of GCS. Hox, Hint, and Hred represent H-proteins bearing covalently attached lipoate (oxidized form), aminomethyllipoate, and dihydrolipoate, respectively.
To gain insights into the structural basis of T-protein catalysis, we have determined the crystal structure of Escherichia coli T-protein (ecT) in complex with Hred from E. coli (ecHred) and 5-methytetrahydrofolate (5-CH3-THF), which mimics the ternary complex in the reverse reaction. The structure revealed for the first time how the H-protein-bound lipoyl group is inserted into the active site of the other GCS components. The present and previous structural information also provides detailed explanations for the NKH-causing mutations. Moreover, the distance between the tip of the dihydrolipoyllysine arm and the methylene carbon atom of modeled 5,10-CH2-THF predicts the presence of an intermediary that mediates the interaction of both reactants. Based on the structural observation and mutational analysis, we propose a reaction mechanism underlying the T-protein catalysis.
EXPERIMENTAL PROCEDURES
Protein Preparation and Assembly of the ecT·ecH Complex
We expressed and purified recombinant E. coli P-protein (ecP), H-protein (ecH), ecT, and ecT with C-terminal His6 tag (ecT-His6) as described previously (6, 18). Single-site mutations were introduced into ecT and ecT-His6 using pET3a/ET and pET23b/ET, respectively, as a template for PCR-based site-directed mutagenesis method with Pyrobest DNA polymerase (Takara). The second mutations were introduced into the constructs bearing one mutation in the same way. After the sequence verification, the expression plasmids were introduced into E. coli BL21(DE3)pLysS, and mutant proteins were expressed and purified following the procedure described for wild-type protein. Full lipoylation of recombinant ecH was verified by the altered migration on SDS-PAGE, in which lipoylated H-protein migrates faster than unlipoylated ecH (19). However, a portion of the preparation was found to be the aminomethyllipoylated form (ecHint) by mass spectrometric analysis (data not shown). Therefore, ecH preparations were treated with ecT in the presence of THF to degrade ecHint contaminants. We then treated the ecH preparation with 15 mm tri(2-carboxyethyl)phosphine hydrochloride (Fluka) to reduce the lipoate moiety. ecHint was prepared enzymatically. The reaction mixture (3 ml) containing 20 mm glycine, 14 mg of ecH, 50 mm potassium phosphate buffer (KPB) (pH 7.0), 0.25 mm pyridoxal phosphate, 1 mm dithiothreitol (DTT), and 1.75 mg of ecP was incubated for 2 h at 37 °C, and the resulting ecHint was purified by hydroxyapatite column chromatography (Bio-Rad). To prepare the ecHint sample used in its degradation assay, we employed [2-14C]glycine (18.5 GBq/mol) instead of nonlabeled glycine. Excess ecHred or ecHint was incubated with ecT or ecT mutants at 25 °C for 30 min in 20 mm 4-morpholinoethanesulfonic acid (pH 6.0) containing 2 mm DTT, and the complexes were isolated by gel-filtration chromatography on a HiLoad 16/60 Superdex 200 pg column (GE Healthcare). The complex-forming ability of the ecT mutants with ecH was also examined by gel-filtration chromatography.
Crystallization, Data Collection, and Structure Determination
We obtained two types of crystals by the hanging-drop vapor-diffusion method at 15 °C by mixing the ecT·ecHred complex solution (22–25 mg ml−1) with an equal volume of different reservoir solutions. One was bipyramid-shaped crystals, which turned out to contain only ecHred monomer, grown in drops containing 0.095 m 2-[4-(2-hydroxyethyl)-1-piperazinyl]ethanesulfonic acid (pH 7.5), 0.19 m calcium chloride, 26.6% (v/v) polyethylene glycol 400, and 5% (v/v) glycerol (Hampton Research) as the mother liquid. The crystals appeared after 4–5 days and reached a maximum size within 2 weeks. The other crystals containing ecT·ecHred complex were obtained in a drop containing 0.04 m potassium dihydrogen phosphate, 16% (w/v) polyethylene glycol 8000, and 20% (v/v) glycerol (Hampton Research) as the mother liquid. The initial crystals, which assembled a rose, appeared after 2 weeks and were used for microseeding; the clustered crystals were grown to a maximum size in 3–5 days. The plate-like crystals separated from the cluster gave sufficient diffraction. The crystals were equilibrated in the mother liquid and flash-frozen in liquid nitrogen. The complex crystals bound to 5-CH3-THF were prepared by soaking the native crystals in mother liquid containing 1 mm (6S)-5-CH3-THF (Scharcks Laboratory) at 15 °C for 4 h and flash-frozen in liquid nitrogen. We prepared the crystals of ecT mutant (ecTD97N) in complex with ecHint and soaked it in 5-CH3-THF solution in the same way as ecT·ecHred crystals. Diffraction datasets were collected at 100 K using the SPring-8 beamline BL44XU on an imaging plate detector DIP6040 (MAC Science/Bruker AXS) and processed using HKL2000 (20).
We first solved the structure of ecHred by molecular replacement with MolRep (21) using the structure of peaH (Protein Data Bank (PDB) code 1HPC) as a search model, and refined by iterative cycles of refinement with Refmac5 (22) and model building in Coot (23). We then solved the structure of the ecT·ecHred complex by molecular replacement as above using the refined ecHred structure (PDB code 3AB9) and the structure of ecT (PDB code 1VLO) as search models. The structures of the ecT·ecHred complex bound to 5-CH3-THF and the ecTD97N·ecHint complex crystals soaked in 5-CH3-THF were also determined as above using the refined structure of ecT·ecHred as a search model. We prepared molecular graphic figures using PyMol. Supplemental Table S1 summarizes the diffraction and refinement data statistics.
Activity Assays
We determined the catalytic activity of ecT-His6 and its mutants in three different assays. Two of them are the glycine cleavage (forward) and synthesis (reverse) activities in the overall reaction system using limited amount of T-protein coupled with saturated amounts of ecH, ecP, and porcine L-protein (Roche Applied Science) in the presence of folate cofactors. Reactions were conducted according to the published protocol (5, 24) with the following modifications. The reaction mixture for the glycine cleavage reaction contained 10 mm [2-14C]glycine (7.4 GBq/mol), 50 mm KPB (pH 7.2), 0.1 mm pyridoxal phosphate, 0.8 mm THF, 1 mm NAD, 2 mm DTT, 900 pmol of ecH, 120 pmol of ecP, 56 pmol of L-protein, and varying amounts of ecT-His6 or its mutants (0.09–14 pmol) in 0.25 ml. The reaction was carried out for 10 min at 37 °C, and 14C-labeled one-carbon unit derived from glycine was trapped as a dimedone adduct and determined as described previously. The reaction mixture for the glycine synthesis contained 50 mm Tris-HCl (pH 8.5), 10 mm DTT, 0.25 mm pyridoxal phosphate, 1.2 mm 5,10-CH2-THF, 1.2 mm formaldehyde, 20 mm NH4Cl, 2 mm NADH, 20 mm NaH14CO3 (7.4 GBq/mol), 900 pmol of ecH, 120 pmol of ecP, 56 pmol of L-protein, and varying amounts of ecT-His6 or its mutants (0.44–28 pmol) in 0.25 ml. The reaction was carried out for 10 min at 37 °C, and the generated [1-14C]glycine was determined. The third method is the direct ecHint cleavage activity in the absence of THF. The reaction mixture contained 40 μm 14C-labeled ecHint, 50 mm KPB (pH 7.2), 1 mm DTT, and 3.5 μm ecT-His6 or its mutants in 0.25 ml. The reaction was carried out for 30 min at 37 °C, and released 14C-labeled one-carbon unit was determined as a dimedone adduct. The reaction proceeded linearly with time up to 30 min and with the concentration of ecT-His6.
Bioinformatic Analysis of T-protein
We collected T-protein (gcvT) homologues by database searches using Hidden Markov Model (HMM) profiles. To construct HMM profiles, we collected known gcvT sequences stored in the KEGG GENES database (Release 51.0) (25) as listed in supplemental Table S2, and these were subjected to multiple alignment with ClustalX 1.83. Four segments corresponding to the underlined ecT sequence in supplemental Fig. S3A were extracted from the multiple alignments and engaged as the training datasets for HMM profile construction using the HMMER package version 2.3.2 with default parameters. The four profiles obtained were subjected to database searches against KEGG GENES with HMMER. To extract gcvT homologues from the database search results, we collected sequences with E-values smaller than 10−10 against all four profiles and removed sequences too long or too short compared with known gcvT sequences. Finally, these sequences together with two additional sequences P49364 (from Pisum sativum (garden pea)) and Q9WY54 (from T. maritima), which are both stored in Swiss-Prot as known gcvT homologues, were again aligned with ClustalX 1.83.
RESULTS
Overall Structures of ecHred and the ecT·ecHred Complex
To crystallize ecT in complex with ecHred, an ecT·ecHred complex with an apparent mass of ∼57 kDa was isolated by gel-filtration chromatography and subjected to crystallization. Depending on the reservoir, crystals containing only the ecHred molecule and those containing the ecT·ecHred complex were obtained. The ecHred and ecT·ecHred structures were determined at 1.7 Å and 2.0 Å resolutions, respectively, as described under “Experimental Procedures.” ecHred crystallized in space group P43212 containing one monomer in an asymmetric unit (supplemental Table S1). The overall structure of ecHred resembles that of peaH, which is 49.2% identical in sequence (supplemental Fig. S1A), showing a hybrid barrel-sandwich structure with the dihydrolipoyllysine arm situated at a hairpin β-motif between β6 and β7 (supplemental Fig. S1B). Structural alignments of the ecHred, two molecules of dihydrolipoate-bearing peaH in an asymmetric unit (PDB code 1DXM), and aminomethyllipoyllysine-bearing peaH (PDB code 1HTP) showed that the root mean square deviation of the Cα atoms ranged from 0.50 Å to 0.55 Å, whereas each lipoyllysine arm positioned differently, indicating its flexibility (supplemental Fig. S1C).
The ecT·ecHred complex crystallized in space group P1 (supplemental Table S1). The crystals contained four ecT molecules (labeled molA–molD) and two ecHred molecules (molE and molF) in an asymmetric unit (supplemental Fig. S2A), forming two heterotrimeric complexes (molCAE and molDBF). Each heterotrimer consisted of an ecT·ecHred heterodimer (molAE and molBF) (Fig. 2A) and an additional ecT molecule (molC and molD). Two ecT molecules in the same heterotrimer were related by a noncrystallographic 2-fold axis with an interface between α4 and β8 (residues 117–140) of each molecule. There was no direct interaction between molC and molE or molD and molF. Because the ecT·ecHred complex eluted as a heterodimer (∼57 kDa) from gel-filtration chromatography, and the molecular mass in aqueous solution was confirmed by dynamic light scattering, crystal packing and the dimer-forming tendency of ecT may have led to such a complex crystal structure. Pairwise structural alignments between the four ecT molecules in the asymmetric unit showed that the root mean square deviation of the Cα atoms ranged from 0.11 Å to 0.32 Å, with an average value of 0.23 Å. Like T-proteins from other organisms, the ecT protomer consists of three domains arranged in a cloverleaf-like structure with a folate-binding central cavity (Fig. 2A). Minor conformational changes were observed in the backbone conformation of the three regions (regions 1–3) in domain 3 upon ecHred binding (supplemental Fig. S2B). Regions 1 and 2 contain residues contributing to the ecT·ecHred interface as described below. When the ecT·ecHred complex crystals were soaked in the 5-CH3-THF solution, they showed the clear Fo − Fc electron density for 5-CH3-THF in the central cavity of all ecT protomers (data not shown). The structures of two ecHred molecules in the asymmetric unit were nearly identical, giving a root mean square deviation of 0.30 Å between the 112 matched Cα atoms.
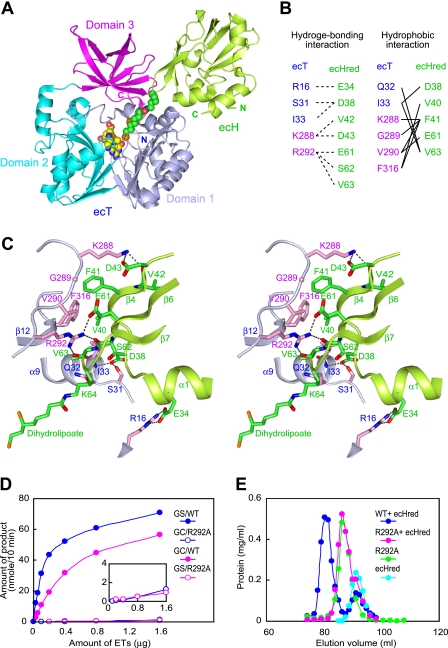
FIGURE 2.
Interface structure of the heterodimer of the ecT·ecHred complex. A, schematic representation of the ecT·ecHred heterodimer (MolAE). Domains 1, 2, and 3 of ecT are colored in light blue, cyan, and magenta, respectively, and ecHred is shown in green with the dihydrolipoyllysine arm shown in CPK model colored in green (carbon), blue (nitrogen), red (oxygen), and orange (sulfur). The bound 5-CH3-THF is also shown in a CPK model colored in yellow (carbon) and the same colors for other atoms as for dihydrolipoyllysine. B, residues contributing to the ecT and ecHred interface. Hydrogen bonding and hydrophobic interactions are summarized. C, close-up view of the interface of molAE in stereo. Key residues from ecT (molA) and ecHred (molE) are shown as sticks with carbon atoms colored in light blue and pink for ecT and green for ecHred. The dihydrolipoyllysine arm is also in stick colored as in A. Hydrogen bonds are depicted as broken lines. D, glycine cleavage (GC) and synthesis (GS) activities of wild-type (WT) and R292A mutant ecT in the overall reaction assays. The inset depicts the trace activities by a large amount of the mutant. E, evaluation of the heterodimer-forming ability of ecT mutant. ecT or ecTR292A was mixed with 1.5–2-fold molar excess of ecHred and subjected to gel filtration. ecT was eluted at the same position of ecTR292A.
Heterodimer Interface of the ecT·ecHred Complex
ecHred binds to the boundary between domains 1 and 3 of ecT, forming an interface with the burial of ∼833.5 Å2 and 845.4 Å2 of exposed surface areas for molAE and molBF, respectively (Fig. 2A) (26). The interface is dominated by hydrogen bonds, which are buttressed by additional van der Waals contacts. Highly conserved residues in both proteins mediated these interactions (Fig. 2B and supplemental Figs. S1A and S3A), although some differences in hydrogen-bonding partners were observed between the two heterodimers (Fig. 2C and supplemental Fig. S2C). A close examination of the interface of the molAE heterodimer revealed four hydrogen bond networks (Fig. 2C). At the center of the interface, the side chain of Arg292 of ecT forms three intermolecular hydrogen bonds to the main-chain carbonyl oxygen of Ser62 and Val63 and to the side chain of Glu61 in ecHred. In the periphery, the side chain of Lys288 of ecT forms hydrogen bonds to the side chain of Asp43 and the main-chain carbonyl oxygen of Val42 in ecHred. Furthermore, the side chain of Asp38 in ecHred is hydrogen-bonded to the side chain of Ser31 and the main-chain amide nitrogen of Ile33 in ecT, and the side chain of Arg16 in ecT to the side chain of Glu34 in ecHred. These hydrogen bonds are stabilized by a patch of van der Waals contacts between residues Gln32, Ile33, Lys288, Gly289, Val290, and Phe316 in ecT and residues Asp38, Val40, Phe41, Glu61, and Val63 in ecHred. The close proximity between Lys288 of ecT and Asp43 of ecH had already been shown in our previous experiment using a zero-length cross-linker (5). The extensive interactions around Arg292 seem to cause a shift of β7 and the hairpin β-motif in ecHred toward the interface (supplemental Fig. S2D). This structural change may force the release of the aminomethyllipoyllysine arm from the binding cleft of aminomethyllipoate-bearing H-protein. The residues in ecHred contributing to the interface correspond very well with those in peaH showing chemical shifts upon pea T-protein binding detected by two-dimensional NMR spectroscopy (17).
Arg292 is one of the residues strictly conserved among T-proteins from 136 organisms ranging from archaebacteria to mammals (supplemental Fig. S3), and its side chain was reoriented to the interface upon ecHred binding (supplemental Fig. S2C). To demonstrate the importance of Arg292 in the protein-protein interaction, we performed a mutational analysis of the residue. Although the behavior of the R292A mutant during expression and purification was similar to that of the wild-type protein, suggesting that the mutation did not affect the overall structure, the mutation abolished not only the glycine cleavage and synthesis activities but also the heterodimer-forming ability (Fig. 2, D and E). When a large amount of R292A was employed in the assay, enzyme activities were observed at trace levels, suggesting that the mutation impairs only the interaction with ecH but not the overall structure (Fig. 2D, inset). These results established the importance of Arg292 in the protein-protein interaction.
Active Site of the ecT·ecHred Complex
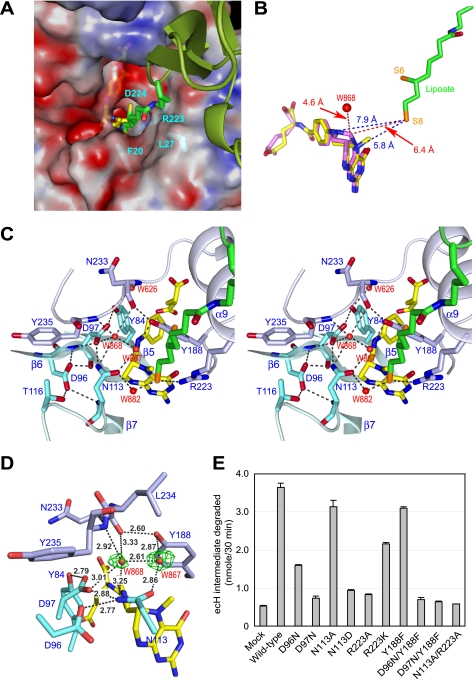
In the complex structure, the dihydrolipoyllysine arm of ecHred is recruited to the active site pocket of ecT with the extending carbon atom chain along with the channel leading to the active site (Fig. 3A). The apparent but not strong omit Fo − Fc electron density for the arm was observed (supplemental Fig. S4A). The arm was coordinated by hydrogen bonds mediated by water molecules from the Nζ atom to Gln32 and Gly221 of ecT and from the S8 atom to Asn113 of ecT. The S8 atom also formed a hydrogen bond to the side chain of Arg223. Extensive van der Waals contacts between the carbon chain atoms and the residues Phe20, Leu27, Arg223, and Asp224 of ecT buttressed the conformation (supplemental Fig. S4B). Because the arm of the ecHred monomer is highly flexible (supplemental Fig. S1B), the arm can spontaneously bind to the active site pocket of ecT upon complex formation. The distances between the S8 atom of the dihydrolipoyllysine arm and the N5, N10, and 5-methyl carbon atoms of 5-CH3-THF were estimated to be 6.0 Å, 7.9 Å, and 4.6 Å, respectively, in the molAE structure (Fig. 3B). These unexpected long distances imply that the direct interaction between the S8 atom and the methylene group of 5,10-CH2-THF is improbable and that some intermediary should mediate the transfer of the methylene group of the folate to the S8 atom in the reverse reaction of T-protein catalysis. We built the structure of 5,10-CH2-THF based on the structure of 5-CH3-THF with Coot, energy-minimized using the Prodrg2 server, and aligned it on 5-CH3-THF in molAE structure (Fig. 3B). The distance between the S8 atom and the C11 atom of modeled 5,10-CH2-THF is estimated to be 6.4 Å, which is still too far for the direct interaction. Although the structure of tmT has been reported as a complex with free dihydrolipoate (11), the direct comparison with the present structure has little meaning; the lipoate derivative depicted in the study was an inactive S-(−)-lipoic acid enantiomer.
FIGURE 3.
Active site of ecT in complex with ecHred and 5-CH3-THF. A, molecular surface of the active site pocket of ecT with the dihydrolipoyllysine arm inserted. Red and blue surfaces indicate negatively and positively charged areas, respectively. ecH is shown schematically, colored in green, and the dihydrolipoyllysine and 5-CH3-THF are in stick colored as in Fig. 1A. The residues of ecT interacting with the dihydrolipoyllysine arm were shown in stick with labels. B, distances between the tip of dihydrolipoate and the atoms of folate cofactors. 5,10-CH2-THF (magenta) was modeled based on the structure of 5-CH3-THF (yellow). Distances (Å) are shown with broken lines. C, close-up view of the active site in stereo. Key residues and four water molecules are depicted in stick (cyan and light blue) and sphere (red), respectively, with labels. The dihydrolipoyllysine and 5-CH3-THF are as in A, and hydrogen bonds are drawn as broken lines. D, putative ammonium-binding site in the reverse reaction. Omit Fo − Fc electron density for the water molecules at the active site of molAE is shown in green, contoured at 2.0σ. Well ordered W868 may occupy the ammonium-binding site. E, ecHint cleavage activity of wild-type and mutant ecTs in the absence of THF. The reaction was carried out as described under “Experimental Procedures.” Mock means nonenzymatical degradation of ecHint. Error bars show variability among three independent measurements.
On one side of the active site pocket, a hydrogen bond network is formed between the side chains of Tyr84, Asp96, Asp97, and Asn113 in ecT. Three water molecules (W868, W867, and W882) participate in further extension of the network. These water molecules are highly ordered and give clear Fo − Fc electron density (Fig. 3, C and D). This hydrogen bond network surrounds 5,10-CH2-THF placing the W868 molecule at a distance of 4.6 Å from the C11 atom of the folate (Fig. 3B). Based on this topology, we propose that the W868-binding site might represent the ammonium-binding site in the reverse reaction. Such an ammonium-binding site has been reported for NAD+ synthetase (27). The coordination observed for W868 is approximately tetrahedral, as expected for an ammonium ion. Water molecules W868 and W867 were retained in all ecT molecules, whereas the position of W882 was replaced by phosphate ion in molC and molD. Our attempts to determine the structure of the aminomethyl moiety and the interacting residues failed; no electron density was observed in the final 2Fo − Fc map around the active site in crystals obtained with an enzymatically prepared ecHint complexed with ecT.
Mutational Analysis of the Active Site Residues
The present structural analysis predicted Asp96, Asp97, Asn113, Tyr188, and Arg223 of ecT as the potential catalytic residues. To test the hypothesis, these residues were mutated; single-site mutants (D96N, D97N, N113D, N113A, Y188F, R223A, and R223K) and double-sites mutants (D96N/Y188F, D97N/Y188F, and N113A/R223A) were expressed with a C-terminal His6 tag to facilitate separation from host T-protein, and purified by affinity chromatography. All mutants behaved similarly to wild-type ecT with a C-terminal His6 tag during expression and purification. All the mutations except for Y188F abolished both the glycine cleavage and synthesis activities (supplemental Fig. S5, A and B). Y188F mutation led to reduction of the activities by 80%. All mutants retained their heterodimer-forming ability (supplemental Fig. S5C). In addition, we examined the direct ecHint degradation activity in the absence of THF of these mutants. About 4% of the added ecHint was degraded nonenzymatically during the incubation, whereas 31% of the input substrate was degraded by wild-type ecT, giving a turnover number of 0.12 min−1, which was approximately 17,000-fold lower than the value obtained for the overall glycine cleavage reaction in the presence of THF. Interestingly, various mutants showed different degradation activities. Three single mutants (D97N, N113D, and R223A) and three double mutants (D96N/Y188F, D97N/Y188F, and N113A/R223A) showed markedly reduced activities (Fig. 3E). It is noteworthy that single mutants D96N and Y188F retained activities of 34 and 83%, respectively, whereas the double mutation in both residues almost abolished the activity, suggesting the compensatory role of these two residues.
These results encouraged us to prepare crystals comprising ecT mutants and ecHint to determine the structure of the aminomethyllipoyllysine in the active site pocket. Our initial attempts to determine the structure of the aminomethyl moiety and the interacting residues failed; no electron density was observed in the final 2Fo − Fc map around the active site in crystals obtained with an enzymatically prepared ecHint complexed with the wild-type ecT. D96N, D97N, and N113D single mutants without a C-terminal His tag subjected to crystallization with ecHint yielded crystals under the same conditions with ecT·ecHred. Although the ecTD97N·ecHint crystals soaked in 5-CH3-THF gave sufficient diffraction, the final 2Fo − Fc electron density for the active site was the same as ecT·ecHred, indicating the degradation of the aminomethyl moiety during crystallization. Interestingly, the electron density corresponding to 5-CH3-THF was not observed in the crystal, suggesting that Asp97 plays an important role for the binding of folate cofactors.
DISCUSSION
The present structural information provides plausible explanations for the disease-related mutations, which can be divided into three groups. One group of residues has already been shown to be involved in the folate cofactor binding (4). To this group, we now can add Tyr188 of ecT (Tyr197 in mature huT) (Fig. 4, A and B); a Y197C mutation was found in a later onset patient having the heterozygous V184A mutation. As mentioned above, Tyr188 contributes to the hydrogen bond network facing the active site cavity (Fig. 3C). It also has extensive hydrophobic contacts with 5-CH3-THF (supplemental Fig. S6B), and the Y188F mutation severely affected the overall activity (80% reduction) without much affecting the ecHint degradation activity (∼20% reduction, Fig. 3E), suggesting a role in the THF binding.
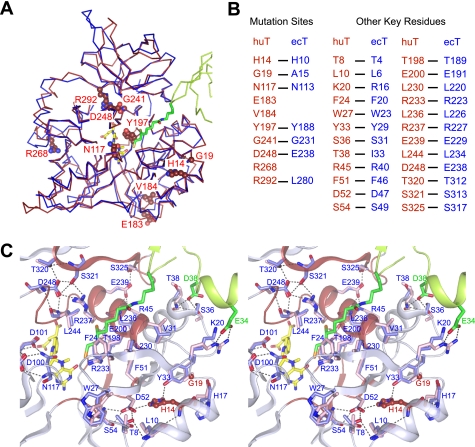
FIGURE 4.
Structural comparison of huT with ecT. A, NKH-related mutation sites mapped on the overall topology of superimposed huT and ecT. The structure of huT (PDB code 1WSV, red) is overlaid on that of ecT (blue) of the ecT·ecHred complex and represented in ribbon. Mutation residues of huT and the corresponding residues of ecT are shown in CPK and stick, respectively, with numbers for huT. ecHred is shown in ribbon colored in green with dihydrolipoyllysine in stick. 5-CH3-THF is shown in stick colored in yellow. B, alignment of key residues of huT and ecT. C, close-up view of the hydrogen bond networks assembling four highly conserved regions in T-protein. Key residues of huT (light blue) and ecT (pink) contributing to the assembly are represented in stick with numbers for huT. Mutant residues are depicted in ball-and-stick representation with atoms colored in red (carbon and oxygen) and blue (nitrogen) with red labels. Four highly conserved regions in the main chain of huT represented schematically (residues 51–56, 196–204, 229–248, and 229–328) are colored in red. The structure of ecHred of the complex and its residues are shown in schematically and stick, respectively, colored in green. Hydrogen bonds are drawn in broken lines.
The present structure revealed the second group of mutations that is involved directly in the catalysis. Asn113 of ecT corresponds to Asn117 in huT, one of the NKH-related residues (Fig. 4, A and B). As will be discussed below, Asn113 plays a central role in T-protein catalysis. Asn113 is almost completely conserved among T-proteins from archaebacteria to mammals except for a group of archaebacteria (group II in supplemental Fig. S3B), in which Asn is replaced with Asp (supplemental Fig. S3A). Structural comparison of ecT with Pyrococcus horikoshii T-protein (a member of group II, PDB code 1V5V) completely overlay Asn113 of ecT on the corresponding Asp116 of P. horikoshii T-protein, suggesting some alternate reaction pathway in these species.
The third group of mutations is found in the N-terminal domain. H14R (His10 in ecT) and G19R (Ala15) mutations were found in NKH patients (Fig. 4A). In the huT structure, the invariant His14 is involved in the hydrogen bond network between Thr8, Leu10, His17, Tyr33, Asp52, and Ser54, which assembles the N-terminal region of T-protein (residues 1–56). With the aid of extensive hydrophobic contacts, these interactions place Lys20 (Arg16 in ecT), Phe24 (Phe20), Ser36 (Ser31), His37 (Asn32), and Thr38 (Ilu33) in positions that interact with H-protein and its lipoyllysine arm (Fig. 4, B and C). Deletion of seven N-terminal residues of ecT led to a 25-fold reduction in the affinity for ecHred without much affecting the affinity for other substrates (5). Furthermore, T4A and L6A mutations in ecT made the protein highly sensitive for trypsinolysis (6). This stacked region may, therefore, serve as a base to assemble four highly conserved regions (residues 51–56, 196–204, 229–248, and 317–328 in huT) to form active site. The assembly is mediated mainly by two hydrogen bond networks, which are buttressed by extensive hydrophobic interactions (Fig. 4C). Recombinant huTs with H14R or G19R mutation were hardly expressed as soluble proteins in E. coli (4), suggesting that the patients with these mutation lack the properly folded active T-protein. Altogether, all of the disease-related mutations are now explained on the solid structural basis.
Our structural observation of the ecT·ecHred heterodimer has provided the first molecular insights into how the H-protein-bound lipoyllysine arm enters the active site pocket of the other GCS component enzymes and participates in the catalysis. Both the protein-protein interaction and the interaction between the lipoyl arm and the T-protein contribute to the complex formation. Because the T-protein binds both Hint and Hred with similar affinities, the structure of the tip of the lipoyl group plays only a limited role in the complex formation. Similar pocket-like structures have been reported for the active sites of P- and L-proteins (12, 15, 16), suggesting that the lipoyllysine arm binds to their active sites in a similar configuration, with the area surrounding the hairpin β-motif of ecH functioning as the interface. The interaction over such a limited area may ensure an easy commutation of the H-protein among three enzymes. Among the residues contributing to the interaction, Arg292 of ecT plays a key role in the complex formation and probably in the release of the aminomethyllipoyllysine arm from the binding cleft of ecHint by weakening the interaction between the aminomethyl moiety and the surrounding environment (17). Despite the capture of the dihydrolipoyllysine arm in the active site, the distances between the S8 atom of the dihydrolipoyllysine arm and the N5, N10, and C11 atoms of the folate cofactor were too long to permit direct interaction. It has been speculated that the aminomethyltransfer reaction from aminomethyllipoate of Hint to THF is initiated by the direct attack of the methylene carbon atom (C9) by the nucleophilic N5 or N10 atoms of THF bound to T-protein accompanying the release of ammonia. The subsequent attack on C9 by the N10 or N5 resulted in the cleavage of the C9–S8 bond (28). This mechanism requires the C9 atom of the aminomethyllipoate arm locating within a covalent-bonding distance (∼1.5 Å) from the nucleophilic nitrogen atoms of THF. The distance observed in the structure, however, suggests the presence of an intermediary mediating the transfer reaction rather than the direct interaction.
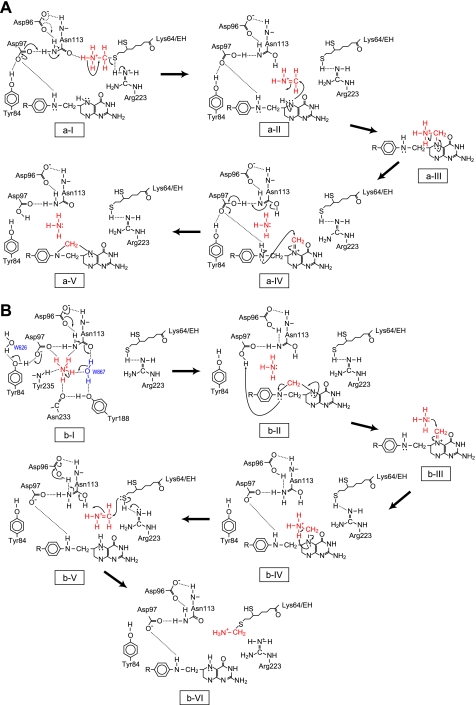
Based on these structural observations together with mutational analyses, we propose a possible mechanism for T-protein catalysis. In the forward reaction (Fig. 5A), the position of W882 is probably occupied by the protonated amino group (ammonium) of the aminomethyllipoyllysine arm, and a hydrogen bond is formed between the N9 atom of the arm and the Oδ atom of Asn113. The initial deprotonation from the ammonium by the Oδ atom induces the cleavage of the C9–S8 bond of the arm with the aid of Arg223 to give a reactive iminium intermediate. Although the Oδ atom of Asn113 itself cannot act as the general base, the electron relay from Asp97 (or Asp96) to Asn113 through the hydrogen bond might confer the nature of a general base on the Oδ atom (step a-I). The released iminium intermediate reacts with the N5 atom of bound THF to give the methylenediamine intermediate (a-II), which undergoes ammonia release (a-III), forming the iminium ion including N5 (a-IV). Subsequent ring closure by the attack of the N10 atom with concomitant deprotonation of N10 by Asp97 results in the formation of 5,10-CH2-THF (a-V). In the reverse reaction (Fig. 5B), an ammonium ion, putatively occupying the position of W868, may be deprotonated by the electron relay-assisted mechanism as above (b-I). The ring breakage of the bound 5,10-CH2-THF with concomitant protonation of the N10 group by Asp97, of which Oδ2 is in near hydrogen-bonding distance, results in an iminium ion including the N5 of the folate (b-II). The attack of ammonia on the iminium carbon forms the N5-bound aminomethyl group (b-III). Subsequent abstraction of the amino-proton by N5 leads to release of the iminium ion (b-IV), which is attacked by the S8 atom of dihydrolipoyllysine with the assistance of Arg223. Finally, aminomethyllipoyllysine is produced (b-VI). The iminium ions are well established intermediates in many reactions, and the detailed reaction mechanism for the nonenzymic condensation of formaldehyde with THF via the iminium ion was reported (29). Furthermore, the electron relay-assisted endowing of the nature of a catalytic base to Asn113 is a mechanism analogous to that reported for LF-transferase (30). Asp97 may play an additional critical role in drawing the folate cofactor into the active site cavity as mentioned above. Asp97 is positioned on the surface of the cavity adjacent to the entrance to the hydrophobic pocket, where the pterin ring is buried (supplemental Fig. S6A). 5-CH3-THF is anchored to the pocket through hydrogen bonds between the 2-amino and 3-imino groups of the pterin ring and the side chain of invariant Glu195 constituting the binding pocket (supplemental Fig. S6B). Asp97 may contribute to drawing the folates by interacting with the 2-amino and 3-imino groups of the pterin ring. Asp97, therefore, seems to be involved in the coordination of the ammonium ion and folate cofactor as well as in the protonation of the N10 atom of 5,10-CH2-THF in the reverse reaction.
FIGURE 5.
Model of the reaction mechanism for T-protein catalysis. The mechanism proposed for the forward (A) and reverse (B) reaction is represented schematically.
Supplementary Material
Acknowledgments
We thank beamline scientists at BL44XU of SPring-8 (Hyogo, Japan) for assistance with data collection, L. Yamada and T. Taniguchi (University of Tokushima) for mass spectrometric analysis, and T. Tsunoda (Tokushima Bunri University) for helpful discussions concerning the reaction mechanism.
This work was supported by grants-in-aid for scientific research (to H. T.) and by the National Project on Protein Structural and Functional Analyses from Ministry of Education, Culture, Sports, Science, and Technology of Japan.
We dedicate this article with our deep condolences to Dr. Yutaro Motokawa who supported this research as a specialist of the glycine cleavage system, and who died during preparation of this paper.
The atomic coordinates and structure factors (codes 3AB9, 3A8J, 3A8I, and 3A8K) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1 and S2.
- GCS
- glycine cleavage system
- THF
- tetrahydrofolate
- 5,10-CH2-THF
- 5,10-methylenetetrahydrofolate
- 5-CH3-THF
- 5-methyltetrahydrofolate
- NKH
- nonketotic hyperglycinemia
- huT
- human T-protein
- Hred
- dihydrolipoate-bearing H-protein
- tmT
- Thermotoga maritime T-protein
- peaH
- pea H-protein
- ecT
- Escherichia coli T-protein
- ecHred
- E. coli Hred
- ecP
- E. coli P-protein
- ecH
- E. coli H-protein
- ecT-His6
- ecT with C-terminal His6 tag
- ecHint
- aminomethyllipoate-bearing ecH
- DTT
- dithiothreitol
- KPB
- potassium phosphate buffer
- PDB
- Protein Data Bank
- HMM
- Hidden Morkov Model.
REFERENCES
- 1.Motokawa Y., Fujiwara K., Okamura-Ikeda K. (1995) in Biothiols in Health and Disease (Packer L., Cadenas E. eds) pp. 389–407, Marcel Dekker, New York [Google Scholar]
- 2.Tada K., Hayasaka K. (1987) Eur. J. Pediatr. 146, 221–227 [DOI] [PubMed] [Google Scholar]
- 3.Nanao K., Okamura-Ikeda K., Motokawa Y., Danks D. M., Baumgartner E. R., Takada G., Hayasaka K. (1994) Hum. Genet. 93, 655–658 [DOI] [PubMed] [Google Scholar]
- 4.Okamura-Ikeda K., Hosaka H., Yoshimura M., Yamashita E., Toma S., Nakagawa A., Fujiwara K., Motokawa Y., Taniguchi H. (2005) J. Mol. Biol. 351, 1146–1159 [DOI] [PubMed] [Google Scholar]
- 5.Okamura-Ikeda K., Fujiwara K., Motokawa Y. (1999) Eur. J. Biochem. 264, 446–452 [DOI] [PubMed] [Google Scholar]
- 6.Okamura-Ikeda K., Kameoka N., Fujiwara K., Motokawa Y. (2003) J. Biol. Chem. 278, 10067–10072 [DOI] [PubMed] [Google Scholar]
- 7.Okamura-Ikeda K., Fujiwara K., Motokawa Y. (1987) J. Biol. Chem. 262, 6746–6749 [PubMed] [Google Scholar]
- 8.Fujiwara K., Okamura-Ikeda K., Motokawa Y. (1984) J. Biol. Chem. 259, 10664–10668 [PubMed] [Google Scholar]
- 9.Cohen-Addad C., Pares S., Sieker L., Neuburger M., Douce R. (1995) Nature Struct. Biol. 2, 63–68 [DOI] [PubMed] [Google Scholar]
- 10.Lokanath N. K., Kuroishi C., Okazaki N., Kunishima N. (2005) Proteins 58, 769–773 [DOI] [PubMed] [Google Scholar]
- 11.Lee H. H., Kim D. J., Ahn H. J., Ha J. Y., Suh S. W. (2004) J. Biol. Chem. 279, 50514–50523 [DOI] [PubMed] [Google Scholar]
- 12.Nakai T., Nakagawa N., Maoka N., Masui R., Kuramitsu S., Kamiya N. (2005) EMBO J. 24, 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattevi A., Schierbeek A. J., Hol W. G. J. (1991) J. Mol. Biol. 220, 975–994 [DOI] [PubMed] [Google Scholar]
- 14.Toyoda T., Suzuki K., Sekiguchi T., Reed L. J., Takenaka A. (1998) J. Biochem. 123, 668–674 [DOI] [PubMed] [Google Scholar]
- 15.Faure M., Bourguignon J., Neuburger M., MacHerel D., Sieker L., Ober R., Kahn R., Cohen-Addad C., Douce R. (2000) Eur. J. Biochem. 267, 2890–2898 [DOI] [PubMed] [Google Scholar]
- 16.Brautigam C. A., Chuang J. L., Tomchick D. R., Machius M., Chuang D. T. (2005) J. Mol. Biol. 350, 543–552 [DOI] [PubMed] [Google Scholar]
- 17.Guilhaudis L., Simorre J.-P., Blackledge M., Marion D., Gans P., Neuburger M., Douce R. (2000) Biochemistry 39, 4259–4266 [DOI] [PubMed] [Google Scholar]
- 18.Okamura-Ikeda K., Fujiwara K., Motokawa Y. (1999) J. Biol. Chem. 274, 17471–17477 [DOI] [PubMed] [Google Scholar]
- 19.Okamura-Ikeda K., Ohmura Y., Fujiwara K., Motokawa Y. (1993) Eur. J. Biochem. 216, 539–548 [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21.Cowtan K. (1994) Newslett. Protein Crystallogr. 31, 34–38 [Google Scholar]
- 22.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Okamura-Ikeda K., Fujiwara K., Motokawa Y. (1982) J. Biol. Chem. 257, 135–139 [PubMed] [Google Scholar]
- 25.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. (2008) Nucleic Acids Res. 36, D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 27.Rizzi M., Bolognesi M., Coda A. (1998) Structure 6, 1129–1140 [DOI] [PubMed] [Google Scholar]
- 28.Douce R., Bourguignon J., Neuburger M., Rébeillé F. (2001) Trends Plant Sci. 6, 167–176 [DOI] [PubMed] [Google Scholar]
- 29.Kallen R. G., Jencks W. P. (1966) J. Biol. Chem. 24, 5851–5863 [PubMed] [Google Scholar]
- 30.Watanabe K., Toh Y., Suto K., Shimizu Y., Oka N., Wada T., Tomita K. (2007) Nature 449, 867–871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.