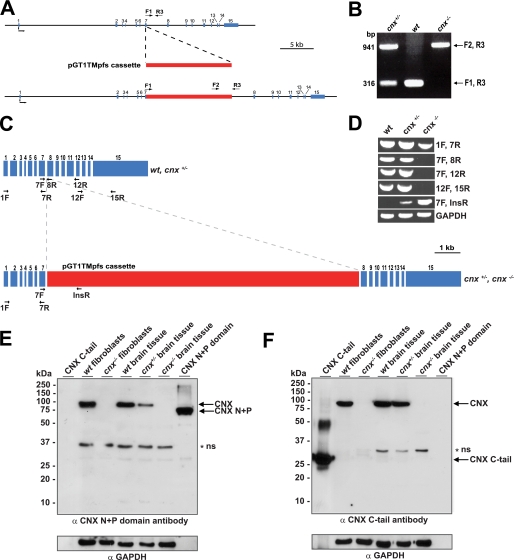
FIGURE 1.
Generation of a calnexin-deficient mouse. A, random gene trapping was used to generate the calnexin-deficient mice. An interruption cassette (pGT1TMpfs cassette, in red) containing β-galactosidase and neomycin genes was inserted into the calnexin gene. The numbers indicate the locations of calnexin gene exons (in blue). Forward (F1 and F2) and reverse (R3) primers are indicated with arrows. B, PCR analysis of genomic DNA isolated from wild-type (wt), heterozygote (cnx+/−), and homozygote (cnx−/−) calnexin-deficient mice. Forward (F1 and F2) and reverse (R3) primers were used as indicated for A. A DNA product of 941 pairs (bp amplified with primers F2 and R3 identifies successful cassette insertion and interruption of the calnexin gene (cnx−/−), whereas a DNA product of 316 bp amplified with primers F1 and R3 indicates the presence of the wild-type allele (wt). The presence of both 941- and 316-bp DNA products identifies heterozygotes. C, schematic representation of calnexin mRNA. The location of the insertion cassette (in red) is shown. The location of specific primers used for RT-PCR analysis in D is indicated in the figure. D, RT-PCR was carried out using wild-type (wt), heterozygote (cnx+/−), and calnexin-deficient (cnx−/−) RNA isolated from brain tissue. Pairs of specific DNA primers used for the analysis are indicated. E and F, Western blot analysis of wild-type, heterozygote, and calnexin-deficient tissues and wild-type and calnexin-deficient fibroblasts with anti-calnexin antibodies. The location of molecular weight markers is indicated to the left of the gel. In E, anti-N terminus (N+P domain) calnexin antibodies were used. In F, the blot was probed with anti-C terminus calnexin antibodies. N+P, N+P domain of calnexin; C-tail, cytoplasmic C-terminal domain of calnexin. The asterisk and ns designate the nonspecific reactive protein band. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.