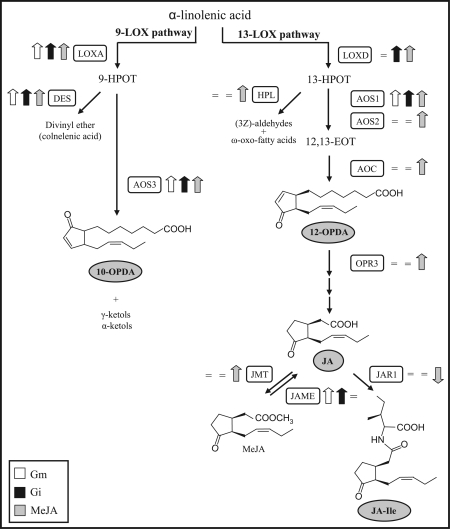
Fig. 3.
Induction of the oxylipin biosynthethic pathway in arbuscular mycorrhizal symbiosis. Metabolic scheme of the oxylipin pathway including the 9-LOX and 13-LOX branches (modified after Wasternack, 2007). Shaded boxes show the metabolites analysed by LC-MS/MS in the present study. Thick arrows indicate the direction of the changes in expression levels compared with non-mycorrhizal control roots (up- or down-regulated) of the genes coding for the enzyme cited, as determined by the transcriptomic analysis; = indicates no changes in gene expression. LOX, lipoxygenase; AOS, allene oxide synthase; AOC, allene oxide cyclase; OPR3, oxo-phytodienoic acid reductase; JMT, jasmonic acid carboxyl methyltransferase; JAME, methyl jasmonate esterase; JAR1, jasmonate-amino synthetase; HPL, hydroperoxide lyase; and DES, divinyl ether synthase.