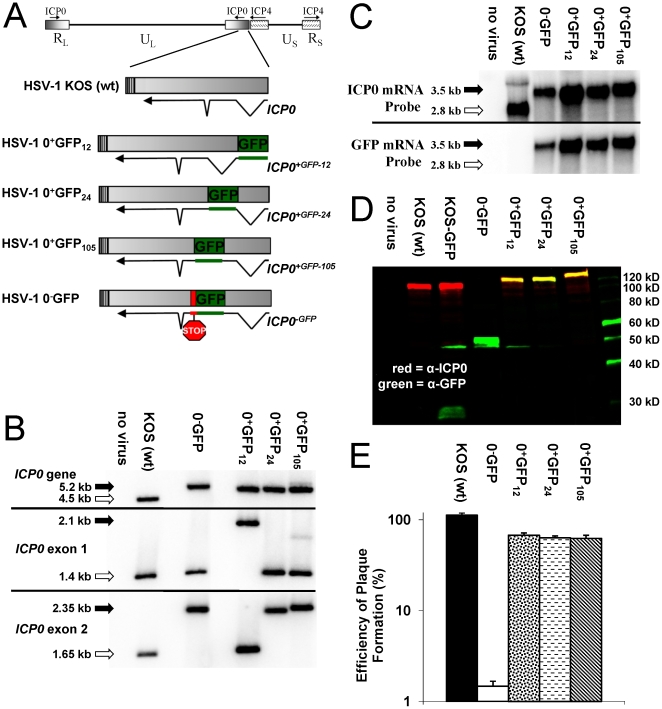
Figure 1. Characterization of HSV-1 viruses that express GFP-tagged ICP0.
(A) Schematic of the wild-type or chimeric ICP0 genes in wild-type HSV-1 strain KOS or the recombinant viruses, HSV-1 0+GFP12, 0+GFP24, and 0+GFP105, and 0−GFP. (B) Southern blot analysis of the ICP0 locus of HSV-1 KOS versus HSV-1 recombinant viruses based on hybridization of an ICP0 intron1-specific probe to DNA digested with StuI and HpaI (ICP0 gene), StuI and BamHI (exon 1 fragment), or PshAI and BssHII (exon 2 fragment). (C) Northern blot analysis with an ICP0- versus GFP-specific probes hybridized to 10 µg total RNA isolated from Vero cells that were uninfected (no virus) or were infected with the specified HSV-1 viruses (MOI = 10; RNA harvested 12 hours p.i.). (D) Two-color Western blot analysis of cells that were uninfected (no virus) or were infected with the specified HSV-1 viruses (MOI = 10; protein harvested 18 hours p.i.). (E) Efficiency of plaque formation by HSV-1 KOS, 0-GFP, 0+GFP12, 0+GFP24, or 0+GFP105 in Vero cells relative to the number of plaques that formed in ICP0-complementing L7 cells (mean ± sem; n = 3 cultures per group).