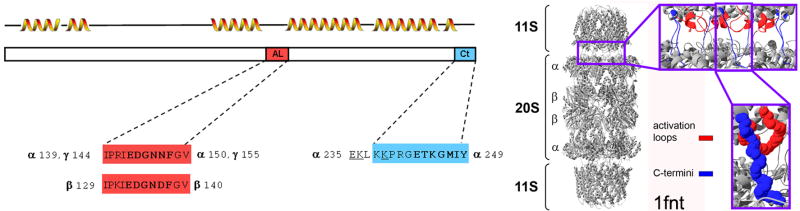
Figure 1.
Design of PA28-derived peptides was based on the crystal structures of PA28α and PA26-20S complex.
Left: The rectangle represents the complete sequence of PA28 subunits. The designed peptides that include the activation loop (AL; bold) are marked in red whereas a peptide incorporating the C terminal sequence (Ct; bold) is marked in blue. The residue positions of peptides specific for each type of subunit are given next to the peptide sequence. The approximate location of helical structures guided by the crystal structure of human PA28α (1avo [11]) is shown above the sequence rectangle. The arrangement of helices and loops is similar in PA28 and PA26 subunits, however the respective AL fragment in PA26 shown in the right panel, extends from residue 96 to 107 (I96PEHKEEDNLGV107).
Right: the positions of AL (red) and Ct (blue) are shown as ribbons (top right) or spacefilled model (bottom right) in the crystal structure of Trypanosoma brucei PA26 (11S; a homolog of PA28) bound to yeast 20S core particle (1fnt [9]). The interface between the activator and the 20S particle is enlarged. Note seven ALs (the leftmost two ALs closely overlap) and seven C-terminal fragments of PA26 heptamer extending toward the top surface of proteasome α ring. A single AL and Ct is further enlarged below. The zoomed-in fragments are marked by purple frames. The ribbon models correspond to secondary structures detected in the crystal structure.