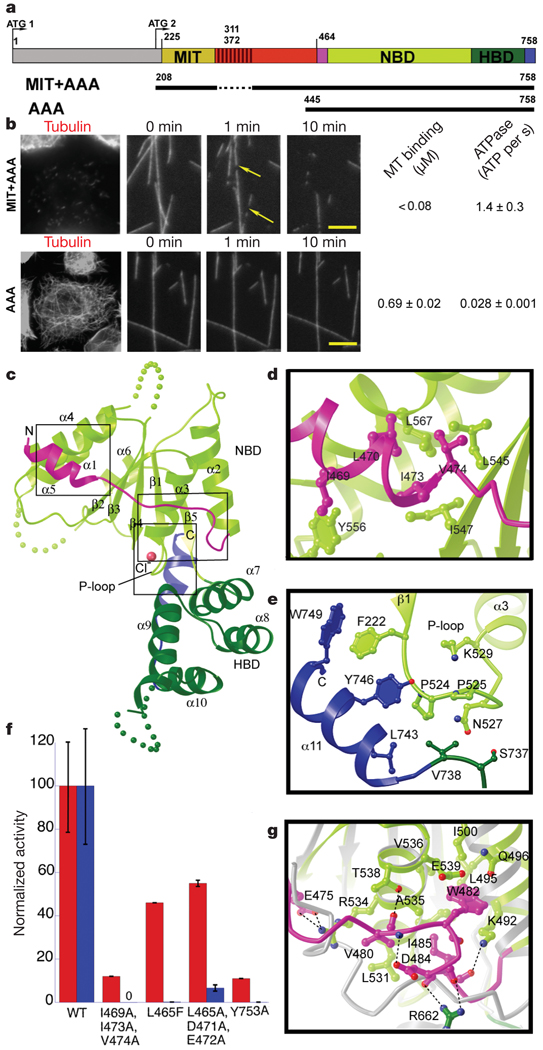
Figure 1. X-ray structure of the nucleotide-free AAA domain of spastin.
a, Domain structure of Drosophila spastin: grey, N-terminal domain; red, linker (exon 4, absent in the shorter isoform of spastin used in this study, is hatched); and the AAA domain (coloured according to the X-ray structure). NBD, nucleotide-binding domain; HBD, four-helix bundle domain. Two potential start codons (ATG) are shown (see Supplementary Methods for discussion). The N-terminal boundary of the AAA domain is based on our X-ray structure and differs from that of ref. 14. A segment of the structurally important N-terminal helix of the AAA domain is within what the authors of ref. 14 define as a microtubule-binding domain. The MIT + AAA and AAA constructs are shown schematically below. b, Left, MIT + AAA disassembles the microtubule network when transfected in Drosophila S2 cells and when added to microtubules in vitro, but AAA has no detectable activity at the same concentration (0.15 µM). (Weak severing is observed at higher concentrations, Supplementary Fig. 1.) Arrows indicate breaks in microtubules. Scale bar, 5 µm. Right, microtubule (MT)-binding and ATPase activities of MIT + AAA and AAA. Microtubule-binding affinity was determined for the Walker B E583Q mutant, which is a stable hexamer and is inactive in severing. c, Ribbon representation of the spastin AAA domain crystal structure. N-terminal helix/loop, magenta; NBD, light green; HBD, dark green; C-terminal helix, blue. The pink sphere depicts a chloride ion. d, Conserved hydrophobic interactions between the N-terminal helix and the main body of the NBD. e, Conserved interactions between the C-terminal helix and the P loop. f, ATPase (red) and microtubule-severing (blue) rates of N- and C-terminal helix mutants. Error bars represent standard errors of the mean (see Methods). WT, wild type. g, Detail of the superposition of spastin and ATP-bound NSF structures15, showing contacts that keep the N-terminal flap of monomeric spastin (magenta) in an open conformation, unable to stabilize the nucleotide or interact with the neighbouring protomer. Spastin is colour-coded as in panel c. NSF is in grey. Dashed lines, hydrogen bonds.