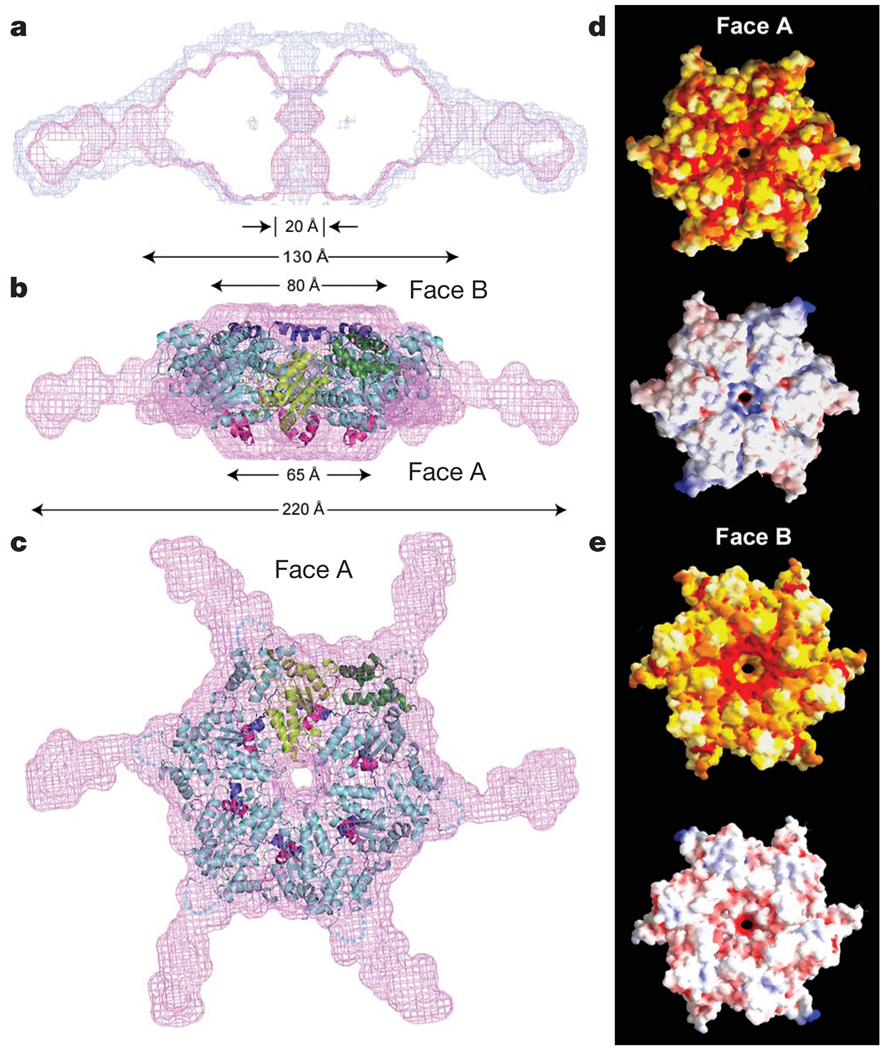
Figure 2. Model of active, hexameric spastin from light and small-angle X-ray scattering.
a, Ab initio SAXS reconstructions21 of C. elegans spastin (MIT + AAA; residues 15–452, ATP-hydrolysis-deficient E278Q mutant). Shown is a cross-section through the filtered (magenta) and composite (blue) SAXS envelopes. The composite structure consists of the aligned, superimposed and summed models from seven independent simulations, whereas the filtered model corresponds to the most probable density map. b, c, Fit of a spastin hexameric model into the SAXS reconstruction (equatorial (b) and axial (c) views). In the absence of an atomic model of the MIT + linker, its precise location within the envelope is uncertain. Maximal diameter is given at various heights of the structure. c shows face A of the hexamer. For details, see Methods. Colour-coding for spastin is as in Fig. 1c. d, e, Surface properties of face A (d) and face B (e). Top image of d and e, solvent-accessible surface of the spastin hexamer model, colour-coded for amino acid similarity as in Supplementary Fig. 3 (white, 40% identity, to dark red, 100% identity, among spastin and katanins). Bottom, solvent-accessible surface of the spastin hexamer model, colour-coded for electrostatic potential (red, negative; blue, positive, ranging from −12 kT to 12 kT).