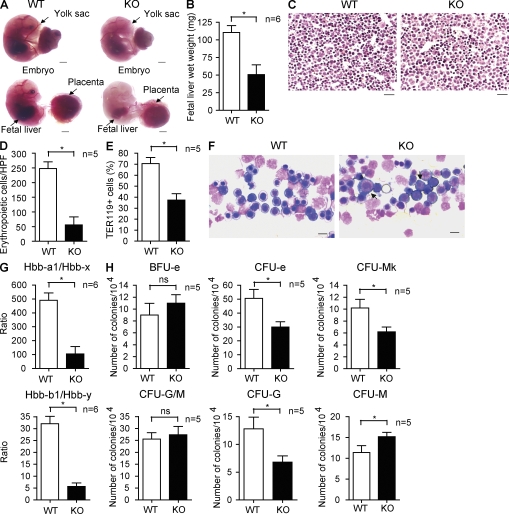
Figure 2.
Erythropoietic defects in fetal liver of SENP1 KO mice. (A) Appearance of WT and SENP1 KO embryos at E16.5. Freshly dissected embryos without staining were photographed. Embryo with yolk sac (top) and without yolk sac (bottom) are shown. Yolk sac, placenta, and fetal liver are indicated. Bars, 200 µM. (B) Wet weight of fetal livers in WT and SENP1 KO embryos (n = 6 each group). Data are mean ± SEM from five fetal livers per group. *, P < 0.01. (C) Histology examination of WT and SENP1 KO E13.5 fetal livers by hematoxylin/eosin staining. Bars, 100 µM. (D) Number of erythropoietic cells per high-power field (40×) was quantified from five sections. *, P < 0.01. (E) Freshly isolated E13.5 WT or SENP1 KO fetal liver cells were incubated in PharmLyse to remove mature red blood cells, and the remaining erythroid progenitor cells were doubly stained with anti-Ter119 followed by flow cytometry (FACS) analysis. The percentage of Ter119+ cells is shown. Data are mean ± SEM from five fetal livers per group. *, P < 0.01. (F) E13.5 fetal liver touch preparations and May-Giemsa staining. Arrows indicate proerythroblasts. Bars, 20 µM. (G) Quantitative RT-PCR analysis of hemoglobin (Hbb) forms in E13.5 WT and SENP1 KO fetal livers. 18S rRNA was used for normalization. Hbb-a1 and Hbb-b1 are adult hemoglobin, whereas Hbb-x and Hbb-y are the corresponding embryonic forms. Data are mean ± SEM from six embryos of each genotype. *, P < 0.01. (H) E13.5 fetal liver cells from WT and SENP1 KO mice were used for colony forming assays of immature BFU-e, CFU-e, CFU-mk, CFU-G/M, and CFU-G or CFU-M. 104 cells were seeded as duplicates from WT and SENP1 KO fetal livers (n = 5 each). *, P < 0.01. Data are mean ± SEM from triplicates of two independent experiments.