Fig. 2.
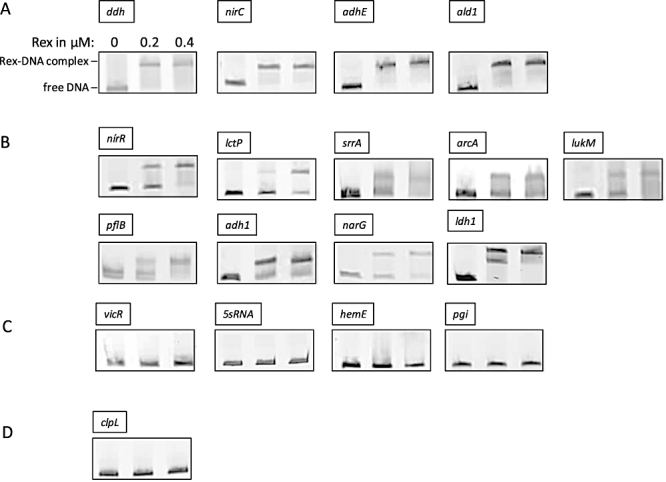
Verification of Rex binding sites by electrophoretic mobility shift assays (EMSAs). Binding of purified Rex to the promoter regions of different genes using EMSAs. The resulting protein–DNA complexes were separated from unbound DNA fragments using native polyacrylamide gels. The DNA fragments were visualized by ethidium bromide staining. Formation of stable Rex–DNA complexes resulted in one or more distinct shifted DNA bands. DNA fragments containing the putative Rex binding site with different Rex binding affinities are shown: (A) strong binding affinity indicated by a complete shift in the presence of 0.2 µM Rex protein; (B) low binding affinity indicated by an incomplete shift in the presence of 0.2 µM Rex protein; (C) no binding in the presence of 0.2 and 0.4 µM Rex protein. (D) As a negative control the upstream region of the anaerobically induced clpL gene which lacks a putative Rex binding site was used.
