Table 2.
Rex-regulated proteins in S. aureus.
| Ratio 30 (60) minutes after oxygen limitatione |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus (8325)a | Proteinb | ROPc | Spotd | 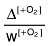 |
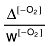 |
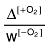 |
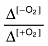 |
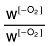 |
Locus (COL)f | Description (COL)f |
| Class I proteins | ||||||||||
| SAOUHSC_00206 | Ldh1 | + | 1 | 13.4 (18.6) | 1.5 (2.8) | 1.7 (5.5) | 0.9 (0.5) | 7.7 (3.4) | SACOL0222 | l-lactate dehydrogenase |
| 2 | 16.2 (12.5) | 3.9 (2.5) | 6 (6.6) | 0.6 (0.4) | 2.7 (1.9) | SACOL0222 | ||||
| 3 | 3.5 (5.5) | 1.3 (1) | 2.2 (2.5) | 0.6 (0.4) | 1.6 (2.2) | SACOL0222 | ||||
| 4 | 8.9 (10.3) | 2 (1.4) | 3.7 (5.9) | 0.5 (0.2) | 2.4 (1.8) | SACOL0222 | ||||
| SAOUHSC_01586 | SrrA | + | 2.9 (3.1) | 1.4 (1.2) | 1 (1.6) | 1.4 (0.8) | 2.9 (2) | SACOL1535 | DNA-binding response regulator | |
| Class II proteins | ||||||||||
| SAOUHSC_00113 | AdhE | + | 1 | 13.4 (6.9) | 48.1 (12.4) | 7.9 (1.8) | 6.1 (7.1) | 1.7 (4) | SACOL0135 | Alcohol dehydrogenase, iron-containing |
| 2 | 14.6 (12.3) | 67.8 (17.8) | 8.9 (2.8) | 7.6 (6.4) | 1.6 (4.4) | SACOL0135 | ||||
| 3 | 3.9 (3.4) | 21.2 (5.3) | 2.9 (0.6) | 7.4 (9.1) | 1.4 (5.7) | SACOL0135 | ||||
| SAOUHSC_00187 | PflB | + | 1 | 1 (0.7) | 9.2 (3) | 0.5 (0.6) | 16.9 (5.5) | 1.9 (1.3) | SACOL0204 | Formate acetyltransferase |
| 3 | 1.2 (0.7) | 9.3 (3.9) | 0.3 (0.2) | 28.2 (20) | 3.6 (3.7) | SACOL0204 | ||||
| 4 | 1.2 (1.1) | 7 (3.6) | 0.1 (0.1) | 72.8 (39.6) | 12.4 (11.7) | SACOL0204 | ||||
| 5 | 0.9 (0.5) | 5.1 (2.6) | 0.1 (0.1) | 43.2 (43.1) | 7.9 (8.1) | SACOL0204 | ||||
| SAOUHSC_00608 | Adh1 | + | 2.5 (5.2) | 9.1 (3.2) | 0.8 (0.7) | 11 (4.4) | 3 (7.2) | SACOL0660 | Alcohol dehydrogenase | |
| Class III proteins | ||||||||||
| SAOUHSC_00375 | GuaA | 1 (1.6) | 2.4 (4.9) | 1.5 (1.9) | 1.6 (2.7) | 0.6 (0.9) | SACOL0461 | GMP synthase/glutamine amidotransferase protein | ||
| SAOUHSC_00535 | – | 1.3 (1.6) | 3.3 (2.2) | 1.1 (1.5) | 2.9 (1.5) | 1.2 (1.1) | SACOL0599 | Hypothetical protein | ||
| SAOUHSC_00909 | – | 0.8 (0.8) | 3.4 (1.4) | 0.5 (0.5) | 6.6 (3.1) | 1.6 (1.7) | SACOL0976 | Hydrolase, haloacid dehalogenase-like family | ||
| SAOUHSC_01064 | Pyc | 2 | 1.4 (11.4) | 1.7 (1.4) | 5.3 (6.8) | 0.3 (0.2) | 0.3 (1.7) | SACOL1123 | Pyruvate carboxylase | |
| SAOUHSC_01218 | SucD | 1.4 (1.1) | 3.8 (1.5) | 1.6 (1.2) | 2.4 (1.3) | 0.9 (0.9) | SACOL1263 | Succinyl-CoA synthetase subunit alpha | ||
| SAOUHSC_01266 | – | 1.3 (1.2) | 5.5 (4.4) | 6.5 (5.2) | 0.8 (0.8) | 0.2 (0.2) | SACOL1308 | Pyruvate ferredoxin oxidoreductase, alpha subunit | ||
| SAOUHSC_01337 | Tkt | 1.8 (1.8) | 2.6 (3.4) | 1.9 (2.3) | 1.4 (1.4) | 0.9 (0.8) | SACOL1377 | Transketolase | ||
| SAOUHSC_01605 | Gnd | 1.1 (1.5) | 5 (1.7) | 1.1 (0.8) | 4.7 (2) | 1 (1.7) | SACOL1554 | 6-phosphogluconate dehydrogenase | ||
| SAOUHSC_01737 | AspS | 1.4 (1.6) | 2 (3.4) | 2.6 (5.6) | 0.8 (0.6) | 0.5 (0.3) | SACOL1685 | Aspartyl-tRNA synthetase | ||
| SAOUHSC_01839 | TyrS | 1 (1.4) | 1.3 (3.2) | 0.8 (1.8) | 1.6 (1.8) | 1.3 (0.8) | SACOL1778 | Tyrosyl-tRNA synthetase | ||
| SAOUHSC_02142 | AldA2 | 4.5 (1.1) | 0.5 (0.8) | 1.1 (1.1) | 0.4 (0.7) | 4 (1) | SACOL1984 | Aldehyde dehydrogenase | ||
| SAOUHSC_02441 | Asp23 | 1 | 0.8 (0.6) | 3 (2.5) | 0.9 (0.8) | 3.4 (3.3) | 0.9 (0.8) | SACOL2173 | Alkaline shock protein 23 | |
| SAOUHSC_02561 | UreC | 1.9 (1.4) | 2.3 (3) | 2 (1.9) | 1.2 (1.6) | 1 (0.7) | SACOL2282 | Urease subunit alpha | ||
| SAOUHSC_02862 | ClpL | 1.5 (1.3) | 2.9 (4.1) | 0.6 (0.3) | 5.2 (12.8) | 2.6 (4) | SACOL2563 | ATP-dependent Clp protease, putative | ||
| Class IV proteins | ||||||||||
| SAOUHSC_00338 | MetE | 2 | 1.6 (1.4) | 0.3 (0.3) | 1 (1.3) | 0.3 (0.2) | 1.5 (1.1) | SACOL0428 | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | |
| SAOUHSC_00365 | AhpC | 3 | 0.2 (0.8) | 0.4 (0.6) | 0.8 (1.1) | 0.6 (0.6) | 0.2 (0.8) | SACOL0452 | Alkyl hydroperoxide reductase, C subunit | |
| SAOUHSC_00794 | GapR | 0.7 (1) | 0.3 (0.6) | 0.3 (0.3) | 1 (2.2) | 2.8 (3.8) | SACOL0837 | Gap transcriptional regulator | ||
| SAOUHSC_00795 | GapA1 | 1 | 0.6 (0.7) | 0.3 (0.6) | 0.3 (0.1) | 1.1 (5.1) | 2.2 (5.7) | SACOL0838 | Glyceraldehyde 3-phosphate dehydrogenase | |
| 2 | 0.7 (1.3) | 0.2 (0.7) | 0.2 (0.2) | 1.1 (4.1) | 3.4 (7.4) | |||||
| 3 | 1 (0.6) | 0.3 (0.6) | 0.3 (0.1) | 1.1 (5.4) | 3.6 (5.3) | |||||
| SAOUHSC_00796 | Pgk | 2 | 0.8 (1) | 0.3 (0.9) | 0.2 (0.2) | 1.3 (4.7) | 3.6 (5.4) | SACOL0839 | Phosphoglycerate kinase | |
| SAOUHSC_00831 | – | 0.6 (0.6) | 0.2 (0.7) | 0.4 (0.4) | 0.6 (1.5) | 1.3 (1.2) | SACOL0872 | OsmC/Ohr family protein | ||
| SAOUHSC_00920 | FabH | 3 | 1.4 (0.2) | 0.4 (0.3) | 1.3 (0.7) | 0.3 (0.5) | 1 (0.3) | SACOL0987 | 3-oxoacyl-(acyl carrier protein) synthase III | |
| SAOUHSC_01347 | – | 0.2 (0.4) | 1.4 (0.6) | 0.5 (0.9) | 2.7 (0.6) | 0.4 (0.4) | SACOL1385 | Aconitate hydratase | ||
| SAOUHSC_01968 | – | 0.3 (0.3) | 0.3 (0.5) | 0.4 (0.3) | 0.9 (1.5) | 0.9 (0.9) | SACOL1894 | HIT family protein | ||
| SAOUHSC_02244 | – | 0.6 (0.6) | 0.3 (0.4) | 0.7 (0.4) | 0.4 (1) | 0.9 (1.4) | – | |||
| SAOUHSC_02441 | Asp23 | 3 | 0.7 (0.8) | 0.3 (0.5) | 0.6 (0.6) | 0.6 (1) | 1.2 (1.4) | SACOL2173 | Alkaline shock protein 23 | |
| SAOUHSC_02441 | Asp23 | 4 | 0.5 (0.5) | 0.2 (0.6) | 0.3 (0.3) | 0.5 (1.8) | 1.3 (1.6) | |||
Loci are based on TIGR annotation of S. aureus COL (http://cmr.jcvi.org).
Symbols are based on TIGR annotation of S. aureus COL or N315 (italic) using Geneplot of NCBI (http://www.ncbi.nlm.nih.gov).
The presence of a Rex binding site (ROP) in front of the respective genes indicated by +.
Spots representing the same protein are distinguished by an additional spot number.
Protein spots whose synthesis was significantly induced or repressed at least threefold in the mutant compared with the wild-type.
Loci and product descriptions are based on TIGR annotation of S. aureus COL using Geneplot of NCBI.
