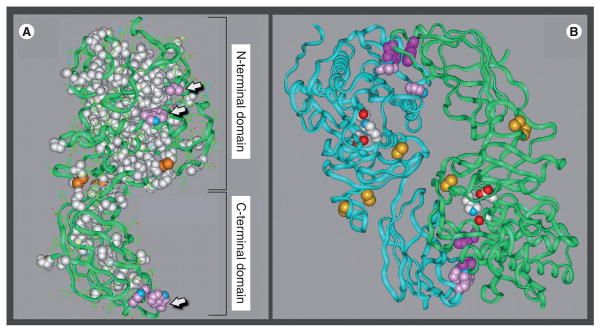
Figure 1. Structural model of hepatic lipase.
(A) The hepatic lipase (HL) monomer is modeled after the crystal structure of pancreatic lipase (PL). Model building and short molecular dynamics runs were performed on a Silicon Graphics Indigo 2 computer with Insight II and Discover 2005 software (Accelrys Inc., CA, USA). The side chains of HL were mapped on to the backbone according to a multiple sequence alignment for HL and PL generated by using the MULTALIN multiple alignment algorithm. The location of the two major folding domains of the HL model is indicated by brackets. Amino acids that are conserved between HL and PL and are depicted in a space-filling form as white; the green ribbons are comprised of residues unique to HL. Also shown are conserved disulfides (orange); arrows point to side chains of surface-located hydrophobic residues (pink). (B) The noncovalent HL homodimer, with the two individual monomers is depicted as ribbons colored blue and green. The side chains of active-site residues are colored red, white and blue; conserved disulfides are highlighted orange; and the side chains of hydrophobic residues comprising putative dimer interaction sites are shown in pink and purple.
Models are courtesy of Keith Munson and Howard Wong.