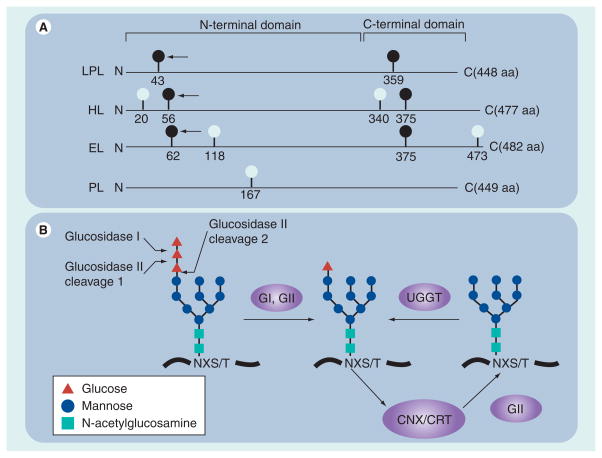
Figure 2. Glycosylation sites and processing events that are important in lipase maturation.
(A) Glycan attachment sites (consensus sequence NXS/T) among members of the human lipase gene family are depicted by light and dark balls. Dark balls represent evolutionarily conserved sites. The arrow points to sites of glycosylation essential in lipase maturation. Also shown are the relative positions of both N- and C-terminal folding domains. (B) Cleavage sites for GI and GII on the unprocessed high mannose chain; this chain is added to NXS/T sites shortly after they emerge from the ribosome during translation elongation. GI and GII cleavages occur rapidly and result in a processed monoglucosylated chain that can bind to CNX or CRT cotranslationally; release occurs after the second cleavage by GII. If the nascent protein becomes misfolded, a single glucose can be added to the unglucosylated chain by the ER lumenal protein, UGGT 1; thus, the lipase can reattach to CNX/CRT in a process termed chaperone cycling.
aa: Amino acid; CNX: Calnexin; CRT: Calreticulin; EL: Endothelial lipase; ER: Endoplasmic reticulum; G: Glucosidase; HL: Hepatic lipase; LPL: Lipoprotein lipase; PL: Pancreatic lipase; UGGT: UDP-glucose: glycoprotein glucosyltransferase.