Abstract
In a 19-year-old severely autistic and mentally retarded girl, a balanced de novo t(14;21)(q21.1;p11.2) translocation was found in addition to a de novo 2.6-Mb 2q31.1 deletion containing 15 protein-encoding genes. To investigate if the translocation might contribute to developmental stagnation at the age of 2 years with later regression of skills, i.e. a more severe phenotype than expected from the 2q31.1 deletion, the epigenetic status and expression of genes proximal and distal to the 14q21.1 breakpoint were investigated in Ebstein Barr Virus-transformed lymphoblast and primary skin fibroblast cells. The 14q21.1 breakpoint was found to be located between a cluster of 7 genes 0.1 Mb upstream, starting with FBXO33, and the single and isolated LRFN5 gene 2.1 Mb downstream. Only expression of LRFN5 appeared to be affected by its novel genomic context. In patient fibroblasts, LRFN5 expression was 10-fold reduced compared to LRFN5 expressed in control fibroblasts. In addition, a relative increase in trimethylated histone H3 lysine 9 (H3K9M3)-associated DNA starting exactly at the translocation breakpoint and going 2.5 Mb beyond the LRFN5 gene was found. At the LRFN5 promoter, there was a distinct peak of trimethylated histone H3 lysine 27 (H3K27M3)-associated DNA in addition to a diminished trimethylated histone H3 lysine 4 (H3K4M3) level. We speculate that dysregulation of LRFN5, a postsynaptic density-associated gene, may contribute to the patient's autism, even though 2 other patients with 14q13.2q21.3 deletions that included LRFN5 were not autistic. More significantly, we have shown that translocations may influence gene expression more than 2 Mb away from the translocation breakpoint.
Key Words: Deletion 2q31.1, Epigenetics, Gene silencing, LRFN5, Translocation
Introduction
De novo and apparently balanced simple reciprocal chromosome translocations are associated with a ∼6% empirical risk for abnormal development, i.e. a risk that is at least twice the background risk [Warburton, 1991; Gardner and Sutherland, 2004]. In about 40% of these abnormal cases, deletion of genes at one or both translocation breakpoints, or a chromosomal imbalance elsewhere in the genome, is found [De Gregori et al., 2007; Baptista et al., 2008; Sismani et al., 2008; Schluth-Bolard et al., 2009]. If no genomic imbalance is detected and no gene is disrupted, the question remains if a de novo translocation should be considered unrelated to an abnormal phenotype. Potentially, translocations and inversions may place genes in a novel genomic context that alters gene expression, either by bringing gene promoters, enhancers or suppressors in the vicinity of other genes (e.g. like the 14;18 translocation increasing MYC expression in acute lymphoblastic leukaemia), by disrupting distal regulatory elements that even may reside within neighbouring genes [Kleinjan and van Heyningen, 2005], or by displacing genes within the nucleus to an environment less favourable for transcription [Cremer and Cremer, 2001]. Translocations affecting R-positive bands (light bands on G-banded karyotypes) are more likely to associate with an abnormal phenotype than translocations through G-positive bands, usually regardless of a gene being disrupted or not [Fantes et al., 2008]. This suggests that the genomic context of the breakpoint has prognostic significance. In quite a number of cases chromosome breaks within 1 Mb upstream or downstream of genes important for normal development have been associated with malformations [Kleinjan and van Heyningen, 2005]. In these cases, disruption of regulatory elements has been the likely mechanism. So far, evidence that balanced translocation may also have adverse effect on gene function due to changes in the epigenetic make-up of genes adjacent to the fusion points, has been lacking.
Here, we present an in-depth study of such a patient: a severely autistic woman with 2 de novo genomic changes, a balanced reciprocal t(14;21)(q21.1;p11.2) translocation and an apparently unrelated 2.6-Mb 2q31.1 microdeletion. Since her severe autism with regression of skills starting at the age of 2 years was not readily explained by the 2q31.1 deletion, we investigated if the balanced translocation might affect expression of nearby genes with potential consequences for her phenotype. We found that expression of LRFN5 was affected by the translocation, even though this gene is 2.1 Mb distal to the 14q21.1 breakpoint. LRFN5 belongs to a family of genes coding for transmembrane proteins that, like the neuroligins, associate with the synapse. The LRFNs are also called synaptic adhesion-like molecules because of their involvement in synaptic development and plasticity [Ko et al., 2006; Morimura et al., 2006].
Patients and Methods
Patient
The index patient is a girl born by caesarean section 10 weeks preterm after amniotic rupture (mother's 4 previous children were also preterm). Her birth weight was 1,200 g, and delivery and neonatal period were described as normal. Written informed consent has been obtained for publication of the patient's pictures and history. At age 4 months, a paediatrician described her as ‘a very alert baby with good eye contact and normal psychomotor development’. Breast feeding was unproblematic, and her growth and development seemed normal until age 6–7 months when her mother first suspected that something was wrong. At that age, she started to appear passive, gave little contact and made few sounds. She was unable to sit without support at age 1 year, walked at age 23 months. She learned to speak a few words (around 50), but most of them were later lost as she became severely autistic, fulfilling the ICD-10 criteria for infantile autism (F84.0). Pictures of the patient, taken at ages 9 months and 6 years, showed no obvious dysmorphism (online suppl. fig. 1; for all online suppl. material see www.karger.com/doi/10.1159/000280290), and on examination at age 16 years she was without dysmorphic signs. She was of short stature (at most 6 cm <2.5th percentile) until age 4, later her stature has been within normal limits (between the 2.5–10th percentile). Her head circumference was initially on the 2.5th percentile and is now on the 25th percentile. Presently, she gives no eye contact, is extremely restless, tears off her clothes, has unmotivated bursts of shouting, and communication with her is impossible. She has ataxia and a broad-based gait, but fine motor skills are better. She needs diapers all the time, has no recognizable sleep pattern and is often awake at night. There is an Angelman syndrome-like fascination for shiny objects and mirrors. In her daily life she needs to be looked after by 1–2 adults all the time to avoid physical destruction of her surroundings.
Formal testing of her intellectual level indicated a loss of skills: At age 4¼ years her developmental age was estimated to be just below 2 years. At age 7 years the general IQ score corresponded to age 1½ years, while the same test done 3 years later gave an IQ score that corresponded to a child aged 1¼ years. On the Vineland Adaptive Behaviour Scale her standard score fell from 32 points at age 7, to 20 points at age 10. On the ADOS-G test (autism diagnostic observation schedule) she scored above threshold for infantile autism on all areas tested.
At age 4 she had an isolated grand-mal seizure. Later her epilepsy has been variable but not problematic or difficult to treat, with absences and partial seizures but few grand-mal seizures. EEGs showed general dysrhythmia. Cerebral CT at age 4 was normal, and so was cerebral MRI at age 11. A metabolic screen was normal. No signs or investigations suggested affected vision or hearing.
A routine cytogenetic test revealed an apparently balanced translocation between chromosomes 14 and 21 with breakpoints in domains 14q21 and 21p11 (online suppl. fig. 2). The latter breakpoint is in the acrocentric p-arm just above the centromere. To investigate if the translocation was really balanced, analysis on the Affymetrix 250K SNP-based GeneChip was done. This revealed no deletions or duplications related to the 14q21 translocation breakpoint, but an apparently unrelated 2.6-Mb 2q31.1 deletion, removing (or interrupting) 15 protein-encoding genes. Parental chromosomes were investigated by G-banding and FISH, and both the balanced 14;21 translocation and the 2q31.1 deletion turned out to be de novo. Her final karyotype was 46,XX,t(14;21)(q21.1;p11.2),ish t(14;21)(RP11-368J16+,RP11-558J21+,RP11-111A21+,RP11-5G7–;RP11-368J16–,RP11-558J21+,RP11-111A21+,RP11-5G7+),arr 2q31.1(172,046,000–174,641,000)×1 dn.
‘Control’ Patients
The first ‘control‘ case is a 12-year-old boy with cleft lip/palate and pes adductus, birth weight 3,790 g (at term). He has facial dysmorphism with short stature, ataxia and choreoathetotic movements. He walked at age 2 years. A major problem has been recurrent bacterial pneumonias. Of note, he is mildly mentally retarded (on WISC-III he scored 3 years below his age level at age 10), has signs of attention deficit and hyperactivity, and is completely without autistic traits. He has a deletion on chromosome 14, karyotype 46,XY,del(14)(q13.1q21.1). This deletion was mapped by 44K oligonucleotide arrays (from Agilent Technologies) to start 33,942 and end 40,834 kb from 14pter (NCBI Build 36), i.e. removing a region of 6.89 Mb containing 36 predicted protein-encoding genes, the first being C14orf147, the last FBXO33.
The second ‘control’ cases are a father and his 1-year-old son with nearly the same karyotype as the above-mentioned control case; 46,XY,del(14)(q13.2q21.3). They both have a 9.17-Mb deletion removing a segment from 34,309 to 43,488 kb from 14pter, including 33 predicted protein-encoding genes; the first being BAZ1A, the last LRFN5. The son was born with hypothyroidism (TSH >500), probably a result of haploinsufficiency of the thyroid transcription factor-1 (TITF1, now called NKX2-1) gene [Devriendt et al., 1998]. In the father hypothyroidism was recently detected, at age 23 years. Other features of NKX2-1 mutations or deletions are neonatal respiratory distress (the father had this) and choreoathetosis [Krude et al., 2002], explaining the choreoathetotic movements in control case 1 and delayed motor development in these patients. The father did not learn to walk before age 4 years, but language development was within normal limits. Similar to the above-mentioned control case the patients are without autistic traits. The father has severe learning difficulties, at most mild mental retardation, and works in a protected environment. The 3.44 Mb larger deletions in these relatives, that also included LRFN5, do not seem to affect the level of mental function compared to control case 1.
Cell Culture and Array-CGH
Epstein Barr virus (EBV)-immortalized lymphoblasts from the patient and a sex-matched blood-donor control were generated according to standard procedures. Primary dermal fibroblasts from the patient and an age- and sex-matched control were grown from skin biopsies, frozen in batches and later used for short-term fibroblast cultures. RNAs from 5 other human primary fibroblast cultures were also used as controls for gene expression. All cells were grown under standard conditions, and total genomic DNA was isolated according to standard procedures. Array CGH investigations (44K arrays, Agilent Technologies) were performed as described previously [Lybaek et al., 2008]. For 250K SNP arrays (Affymetrix), 250 ng of genomic DNA from the patient's lymphoblasts was digested with NspI, ligated to adaptors, amplified with generic primers recognizing these adaptors, and purified. 90 μg of this purified DNA was labelled with biotin, and hybridized in the Affymetrix GeneChip Hybridization Oven 640. Next, the arrays were washed and stained in the Affymetrix GeneChip Fluidics Station 450. Arrays were scanned with the Affymetrix GeneChip Scanner 3000 7G. Samples were required to have a minimum Quality Control SNP call rate of 90% (DM algorithm). To assess copy numbers, samples were analysed with the Copy Number Analyzer for GeneChip (CNAG v2.0) package [Nannya et al., 2005]. The predicted copy numbers were determined using the Hidden Markov Model incorporated in the CNAG package.
Fluorescence in situ Hybridization
The t(14;21) breakpoint was mapped by FISH analysis on methanol/acetic acid fixed lymphoblast and fibroblast cells of the patient, essentially as described previously [de Bruijn et al., 2001]. In short, BAC DNA was biotin-labelled by nick translation, and hybridized to metaphase chromosomes in the presence of a 30-fold excess of Cot 1-DNA. Detection of the hybridizing probe was achieved using fluorescein isothiocyanate (FITC)-conjugated avidin followed by one application step of goat anti-avidin, conjugated with FITC. Pre-labelled reference probes for the centromeres and q-arm telomeres of chromosomes 14 and 21 were obtained commercially (Vysis). A Zeiss Axiophot-2 microscope equipped with the appropriate fluorescence filters was used for analysis of the fluorescence signals. Images were captured by a Leica DC 350 FX camera using the Leica CW 4000 software package.
RNA Extraction and Real-Time Quantitative Reverse Transcription-PCR
Total RNA was isolated from patient and control cells (primary fibroblasts and EBV-transformed lymphoblast cells) using Trizol (Invitrogen), according to the manufacturer's instructions. RNA concentrations were measured using a nanodrop spectrophotometer (Thermo Scientific), and 1 μg of total RNA was reverse transcribed using Omniscript reverse transcriptase (Qiagen) in 20-μl reactions, according to the manufacturer's instructions. This procedure was done twice to obtain independent replicates of each RT reaction. Real-time quantitative RT-PCR (qRT-PCR) was done in duplicate with the SYBR green PCR master mix (Applied Biosystems) using 1 μl of each of the replicate reverse transcriptase reactions. Oligonucleotide primers used to detect expression of the ACTB (β-actin), PNN, FBXO33 and LRFN5 genes were developed using the Vector NTi software package (Invitrogen). Dissociation curve analysis was used to ensure primer specificity and optimal concentration. Expression levels were quantified using the ABI PRISM7500 sequence detection system. The qRT-PCR data was normalized to β-actin, and the average of the 4 replicates was plotted with the standard deviation as error bars.
Chromatin Immunoprecipitation Analysis
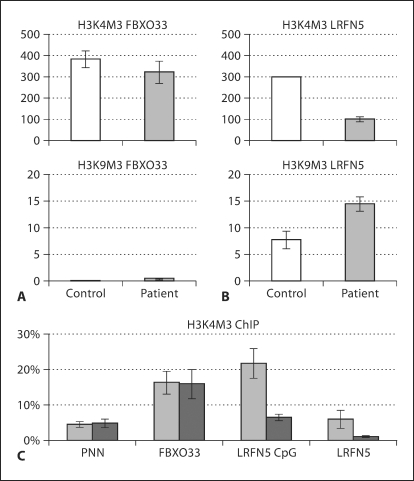
Cultured cells were washed with PBS and cross-linked for 15 min with 1% formaldehyde in minimal culture medium at room temperature. Next, cells were washed with PBS and the chromatin was isolated and sheared by sonication. Protein-DNA complexes were immunoprecipitated as described previously [de Bruijn et al., 2006] using chromatin immunoprecipitation (ChIP)-validated antibodies against histone H3 (histone H3, Abcam ab1791), histone H3 trimethylated at lysine 4 (H3K4M3, Abcam ab8580), histone H3 trimethylated at lysine 27 (H3K27M3, Abcam ab6002), histone H3 dimethylated at lysine 9 (H3K9M2, Abcam ab7312), histone H3 trimethylated at lysine 9 (H3K9M3, Abcam ab8898), and rabbit IgG (as pre-immune control). After ChIP, the formaldehyde-induced crosslinks were reversed and the genomic DNA was recovered. Validation of the ChIP-on-CHIP data (see below) was done with real-time quantitative PCR on non-amplified ChIP samples. The primers used to detect the promoters of the ACTB, PNN, FBXO33 and LRFN5 genes and the LRFN5 upstream CpG island were developed as described above (oligo sequences are given in online suppl. table 1). Quantification and dissociation curve analysis were done using the ABI PRISM7500 sequence detection system. The level of histone modifications was calculated as the average of replicate real-time PCRs, expressed as raw values in arbitrary units (fig. 5A and B) or as percentage of the input (fig. 5C) and plotted with the standard deviation as error bars.
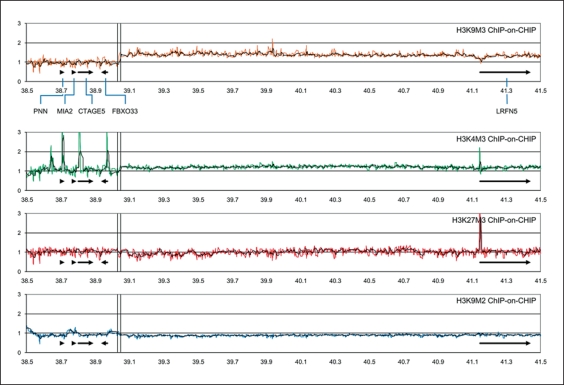
Fig. 5.
Results of real-time PCRs of the PNN, FBXO33 and LRFN5 promoters on H3K4M3 and H3K9M3 ChIP material from control and patient fibroblasts. The FBXO33 promoter-associated levels of H3K4M3 and H3K9M3 were not significantly different between control and patient (A) in contrast to the LRFN5 promoter-associated H3K4M3 and H3K9M3 levels (B). The decreased level of H3K4M3 and increased level of H3K9M3 that was found at the LRFN5 promoter in patient fibroblasts corresponded well with reduced LRFN5 expression (fig. 3). C A more elaborate analysis of H3K4M3 levels at the PNN, FBXO33 and LRFN5 promoters, and the LRFN5-associated CpG island, of control (light grey) and patient (dark grey) fibroblasts. These data demonstrated that the patient-specific decrease of H3K4M3 levels was more widespread within the LRFN5 promoter region. Levels of histone modifications are expressed as arbitrary (raw) values in A and B, or as percentage of the input DNA in C. Bar graphs show average values of independent replicates, with standard deviations as error bars.
High Resolution ChIP-on-CHIP
For ChIP-on-CHIP analysis, 50% of each of 2 replicate ChIP samples (from H3K9M3, H3K4M3, H3K27M3 and H3K9M2 ChIPs) were concentrated using microcon YM-30 spin columns (Millipore) and amplified using the GenomePlex Whole Genome Amplification kit (Sigma). The ChIP-on-CHIP hybridization was done on high resolution NimbleGen HG18 chromosome 13- and 14-specific 385K arrays (C4524-29-01HG18 Tiling Set 29; Roche NimbleGen Systems). This array consisted of 383,527 probes (50-mers), tiling the unique regions of chromosome 13 (95.001–114.127 Mb) and chromosome 14 (18.126–72.700 Mb) uniformly, with a median probe spacing of 100 bp. DNA labelling, array hybridization, post-hybridization washes and scanning were essentially performed according to the manufacturer's instructions (Roche NimbleGen Systems). In short, the ChIP and input (DNA from non-precipitated chromatin) samples were labelled with Cy3- and Cy5-conjugated random nonamers, respectively. The labelled samples were purified, combined, denatured and hybridized overnight at 42°C to the tile-path arrays. After stringent washing the microarrays were scanned using an Axon 4200AL Scanner (Molecular Devices) at 5-μm resolution. The acquired images were analyzed using NimbleScan V2.4 extraction software (Roche NimbleGen Systems), and combined to create pair reports, including raw intensities for each probe and per image. From these data, scaled ratio files were generated, and peaks were defined. For data visualization, the average ratios of 2 replicate experiments were binned per kb and plotted against their chromosomal position. A trend line was added, representing the moving average of 8 subsequent data points.
Results
Fine-Mapping of the Patient's Two de novo Chromosome Aberrations
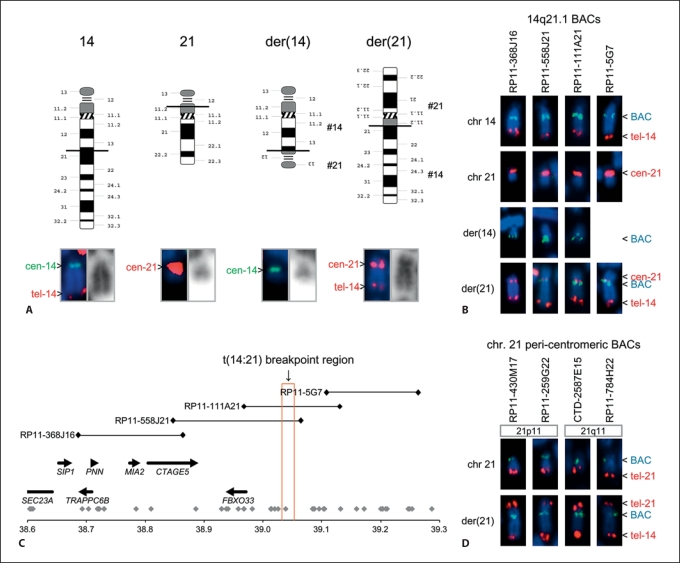
The finding of a de novo apparently balanced 46,XX, t(14;21)(q21.1;p11.2) translocation in a girl with classic infantile autism and severe mental retardation several years ago suggested that the translocation could be related to the phenotype. On the G-banded karyotype, it first seemed that the normal centromere 21 could have been lost and replaced by a neo-centromere at the translocation breakpoint on the large derivative chromosome (online suppl. fig. 2). However, FISH using a centromere 21 alpha-repeat DNA probe gave a weak signal, and a centromere could also be detected by heterochromatin-specific C-banding, indicating that a short alpha-repeat sequence was present (data not shown). Such a short 21 centromere is a common variant in humans, found in around 3.7% of individuals [Lo et al., 1999]. This was later confirmed using centromere 21-flanking BAC probes (fig. 1D). An identical short centromere 21 was found in the father.
Fig. 1.
Translocation breakpoint mapping. A Idiograms (courtesy of David Adler, University of Washington) of the normal and derivative chromosomes 14 and 21 with horizontal black lines indicating the translocation breakpoints. FISH results are shown below to verify the identity of centromeres and telomeres. B Results of BAC-FISH using probes flanking the 14q21.1 breakpoint. C Detailed breakpoint map including the adjacent genes and the position of the BAC-FISH probes. D BAC probes flanking centromere 21 are present on both the p-arm and the q-arm of the derivative chromosome 21, indicating that the centromere 21 is intact and that the breakpoint is in 21p11.
It was initially assumed that the reason for the girl's severe developmental delay was a translocation-related deletion. However, high-resolution chromosome-based HCGH (HR-CGH) and analysis on custom-made 1-Mb resolution BAC arrays indicated that the translocation was balanced (data not shown). To look for mosaicism, chromosome analysis of 30 metaphases from a skin fibroblast culture was also done, with normal result. An alternative explanation was that a gene with critical importance for brain function was disrupted by the translocation, and efforts were taken to map the translocation breakpoints by BAC-FISH, but no gene seemed to be placed within any of the breakpoints.
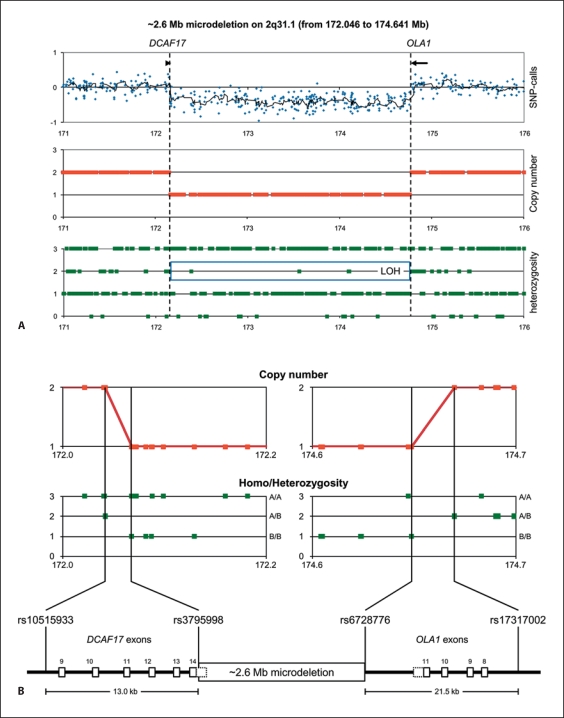
Many years later, patient DNA was examined for copy-number alterations on an Affymetrix 250K GeneChip array, and then a de novo 2.6-Mb 2q31.1 deletion (172.046–174.641 Mb from pter) was revealed (fig. 2). No genomic imbalance was detected at the t(14;21) translocation breakpoints. At this time point, more extensive BAC-FISH breakpoint mapping was also performed, using overlapping BAC probes that provided a complete coverage of the breakpoint area (fig. 1B, C). Of the 4 BAC clones, 2 (RP11-558J21 and RP11-111A21) gave signals on normal chromosome 14, der(14) and der(21), indicating that these were breakpoint-spanning clones. The revised karyotype was 46,XX,t(14;21)(q21.1;p11.2), ish t(14;21)(RP11-368J16+,RP11-558J21+,RP11-111A21+,RP11-5G7–;RP11-368J16–,RP11-558J21+,RP11-111A21+,RP11-5G7+), arr 2q31.1(172,046,000–174,641,000)×1 dn.
Fig. 2.
Affymetrix 250K GeneChip data showing the 2.6-Mb 2q31.1 microdeletion (A), and a detailed map of the deletion breakpoints (B). A Visualization of the microdeletion by raw log2 intensity ratios (blue dots) with a running average (black line) and the CNAG-derived copy numbers (red dots). The microdeletion is further highlighted by a long stretch of loss of heterozygosity (blue square; LOH) in the SNP genotypes (green dots). The proximal and distal breakpoints are indicated as dotted lines. B Detailed map of the microdeletion breakpoint regions and the breakpoint-flanking SNPs. CNAG-derived copy numbers (red dots) and SNP genotypes (green dots) show that the proximal breakpoint is located within a 13.0-kb region encompassing the 3′-end of the uncharacterized DCAF17 gene. The distal breakpoint is located within a 21.5-kb region encompassing the 3′-end of the OLA1 gene. Exons of both genes are shown as numbered boxes, and the 3′UTRs are shown as dotted boxes.
We determined the approximate location of the breakpoint on chromosome 14 from 2 estimates. First, we gauged that BAC RP11-558J21 had 10% of its signal on der(21). Secondly, we estimated that BAC RP11-111A21 had a little more than half (55%) of its signal on der(21). Together with the respective BAC sizes, these estimates pointed to a breakpoint location at 39.042 Mb. Then, to account for errors in our estimates, we defined the breakpoint region between 39.032 and 39.052 Mb from 14pter (position according to NCBI Build 36), i.e. 0.1 Mb distal to FBXO33 (fig. 1C). FBXO33 is the last gene in a 0.4-Mb cluster of 7 genes (SEC23A, SIP1, TRAPPC6B, PNN, MIA2, CTAGE5, and FBXO33) just upstream of the breakpoint. The 14q21.1 breakpoint was also at the beginning of a 5-Mb ‘gene desert’ containing only 1 protein-encoding gene, LRFN5, situated 2.1 Mb distal to the breakpoint. Half a megabase upstream from LRFN5 there is a spliced non-coding RNA (BX248273) with 4 exons but no open reading frame.
The breakpoint on chromosome 21 was located adjacent to or possibly even in the stalk of the acrocentric 21p arm, as illustrated in figure 1. An acrocentric 21p arm with its satellites was fused to the FBXO33 gene cluster by the translocation, creating a bisatellited der(14) chromosome (fig. 1A). Of note, the 21p satellite was always found to be functionally inactive by NOR-banding (data not shown).
Silenced Expression of LRFN5, 2.1 Mb Downstream of the Translocation Breakpoint
Even though the nature of the patient's 2 different de novo chromosome aberrations now was known in detail, it was not straight-forward to explain the development from apparent normality at age 4 months into increasingly severe autism from age 1–2 years and onwards. None of the deleted genes were known to be associated with autism or progressive mental retardation (see Discussion). We therefore wondered if the translocation, albeit balanced and without disrupted genes, also influenced this severe phenotype. The silenced ribosomal RNA genes, located on the acrocentric 21p arm on the der(14) chromosome, might affect the chromatin structure and expression of the FBXO33 cluster 0.1 Mb away. Alternatively, the more proximal part of the 21p arm with the adjacent centromere 21 on the der(21) chromosome might influence LRFN5 expression, despite this gene being 2.1 Mb distal to the translocation breakpoint.
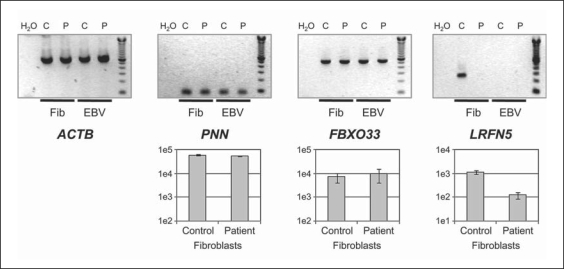
As a first step, we investigated whether gene expression was affected by the translocation. Using RT-PCR, we measured the expression of 2 genes from the upstream gene cluster (PNN and FBXO33) and the downstream gene LRFN5. We found no clear difference in PNN or FBXO33 expression between the patient and age- and sex-matched controls in lymphoblasts or fibroblasts (fig. 3). Expression of the LRFN5 gene was undetectable in both patient and control lymphoblast cells. In fibroblasts, however, LRFN5 expression was clearly detectable in half of the different control cultures (3 of 6), the other half being completely expression-negative. The patient fibroblasts belonged to the LRFN5 expression-positive group, but here the expression level was 10-fold reduced compared to the expression-positive control fibroblasts (fig. 3 and online suppl. fig. 3). These RT-PCRs were subsequently validated by real-time QRT-PCR (fig. 3, bar graphs), and the origin of the PCR products was sequence-verified.
Fig. 3.
Expression analysis of the β-actin control (ACTB) and three 14q21.1 breakpoint-flanking genes on der(14) (PNN and FBXO33) and der(21) (LRFN5) in Epstein-Barr virus transformed lymphoblasts (EBV) and skin fibroblasts (Fib) of control (C) and patient (P). PNN and FBXO33 expression was detected in all samples and did not differ significantly between patient and control cells. LRFN5 expression was readily found in control fibroblasts, but was almost undetectable in patient fibroblasts. Bar graphs show the expression of PNN, FBXO33 and LRFN5 determined by real-time RT-PCR on control and patient fibroblasts. Expression levels are means of 4 replicate PCRs, error bars indicate the standard deviations, the scale is logarithmic.
The Translocation Caused Heterochromatinization of the LRFN5 Locus
To seek an explanation for the apparent LRFN5 downregulation in patient fibroblasts, we employed ChIP using fibroblasts from the patient and an LRFN5 expression-positive control to identify epigenetic changes related to the 14q21.1 breakpoint. The antibodies used were against histone H3, trimethylated lysine 4 of histone H3 (H3K4M3), bi- and trimethylated lysine 9 of histone H3 (H3K9M2 and H3K9M3), and trimethylated lysine 27 of histone H3 (H3K27M3). H3K4M3 is a marker of an open chromatin configuration, found in the promoter regions of active genes (euchromatin), H3K9M2 and H3K27M3 are markers associated with gene silencing, while H3K9M3 is a marker of transcriptional repression, i.e. even more inaccessible chromatin environments (heterochromatin) [Peterson and Laniel, 2004; Kouzarides, 2007]. The precipitated DNA of the patient's fibroblasts was subsequently analyzed on chromosome 14 tile-path oligonucleotide arrays (fig. 4) and validation of these data was performed by real-time quantitative PCR on non-amplified ChIP material from the patient and control fibroblasts (fig. 5), with one exception (online suppl. fig. 4).
Fig. 4.
Results of patient fibroblast ChIP-on-CHIP experiment: Chromatin from patient fibroblast cell lysates was immunoprecipitated by antibodies against K3K4M3, H3K9M2, H3K9M3 and H3K27M3. DNA from these ChIP samples was amplified and hybridized to tile-path chromosome 14 oligonucleotide arrays, using input DNA as the comparative control. The curves show the average ratio of the hybridization signals of 2 independent immunoprecipitates, compared to the input DNA (for details, see the Methods section). The 14q21.1 translocation breakpoint region, the same as in figure 1, is marked with a vertical double line. There is a marked increase in H3K9M3-associated and a smaller increase in H3K4M3-associated DNA starting at the breakpoint and including the LRFN5 gene, and peaking at the LRFN5 promoter for H3K4M3- and H3K27M3-associated DNA.
ChIP-on-CHIP hybridization to tile-path arrays revealed elevated levels of the heterochromatin signature H3K9M3 starting distal to the FBXO33 gene cluster (fig. 4). Remarkably, the change from low to elevated H3K9M3 levels was sudden and coincided exactly with the BAC-FISH mapped t(14;21) breakpoint (fig. 4). This level of H3K9M3 remained elevated throughout the LRFN5 locus, indicating that the entire 5-Mb ‘gene desert’ with LRFN5 in the middle was packed into heterochromatin. The same analysis revealed several peaks of the active-gene signature H3K4M3, both within the upstream FBXO33 gene cluster (associated with the PNN, CTAGE5 and FBXO33 promoters) and at the LRFN5 promoter (fig. 4). Interestingly, we observed elevated levels of H3K4M3 (similar to what was seen for H3K9M3), also starting at the translocation breakpoint and stretching out into the same region, again indicating that this elevation is causally related to the breakpoint (fig. 4). No such putative breakpoint effect was observed in the H3K27M3 and H3K9M2 ChIP-on-CHIP analyses. The H3K27M3 ChIP-on-CHIP analysis gave a single distinct peak at the LRFN5 promoter (fig. 4). This co-occurrence of H3K4M3 and H3K27M3 at the LRFN5 promoter is reminiscent of the so-called bivalent domains [Kouzarides, 2007]. A similar bivalent domain configuration has been found at the LRFN5 promoter in mouse and human embryonic stem cells [Ku et al., 2008].
To validate these patient-derived ChIP-on-CHIP chromatin signatures, and to verify whether these were unique to the patient cells, we performed real-time PCRs for the FBXO33 and LRFN5 promoters on non-amplified ChIP material from patient and control fibroblasts. The chromatin signature of the FBXO33 promoter clearly showed high levels of H3K4M3, and hardly detectable levels of H3K9M3 in fibroblasts of control and patient (fig. 5A). This signature is typical of active genes and this is in full support of our RT-PCR results (fig. 3). In contrast, the LRFN5 promoter had a distinctly different signature, with lower levels of H3K4M3 and increased levels of H3K9M3 in the patient (fig. 5B). This signature is typical for lowly expressed genes, a finding that is also fully supported by our RT-PCR results. The level of H3K9M3 at the LRFN5 promoter was increased about 2-fold in the patient, as compared to the control (fig. 5B). These results validated our ChIP-on-CHIP data and indicated that the increased heterochromatinization (measured by the H3K9M3 levels) of the LRFN5 locus was specific for the patient. Of additional interest, however, was the sharply decreased level of H3K4M3 at the LRFN5 promoter in the patient fibroblasts compared to control fibroblasts (fig. 5B). The higher LRFN5 promoter H3K4M3 level in control fibroblasts is in line with the observed LRFN5 expression in these cells (fig. 3). Further real-time PCRs on replicate H3K4M3 ChIP material from control and patient fibroblasts confirmed these findings, and showed that the patient fibroblast LRFN5 promoter H3K4M3 level was decreased strongly (up to 6-fold compared to the input level) in a wider region that included the CpG island located 1.2 kb upstream (fig. 5C). This result also correlates well with the about 10-fold lower LRFN5 expression level in patient fibroblasts (fig. 3).
Another factor that might play a role is the level of H3K27M3, which appeared as a peak exactly at the LRFN5 promoter in the ChIP-on-CHIP data from patient fibroblasts (fig. 4). Validation of these results by real-time PCR on non-amplified H3K27M3 ChIP material showed that this H3K27M3 signature was not present at the LRFN5 promoter in the control fibroblast cells (data not shown). However, we also found that the H3K27M3 signal in the patient's fibroblasts was less strong than what could be expected from the ChIP-on-CHIP data (fig. 4), indicating that the H3K27M3 peak was partially caused by the genome amplification step prior to array hybridization. To address this, we performed semi-quantitative PCR of the FBXO33 and LRFN5 promoters on the amplified ChIP material (the same that was used for the array hybridizations). In this way we were able to validate the presence of an H3K27M3 peak at the LRFN5 promoter of the patient (online suppl. fig. 4).
Haploinsufficiency of LRFN5 Is Not Associated with Autism
To decipher the clinical significance of the 2 de novo changes we looked for control cases for the 2q31.1 deletion, as well as for the FBXO33 gene cluster and/or LRFN5 haploinsufficiency. Regarding the latter, we picked up 3 patients with relevant chromosome 14 deletions in our diagnostic laboratory: a boy with a 6.89-Mb chromosome 14 deletion that included the FBXO33 cluster but not LRFN5, and a father and son with a quite similar 9.17-Mb deletion that also included LRFN5. All 3 patients were found after array-based testing for genomic copy number aberrations in our routine diagnostic laboratory. They all had a similar degree of cognitive impairment, at most a mild degree of mental retardation, and were completely without autistic traits. Most importantly, inclusion of the LRFN5 locus in the deletion did not appear to affect the phenotype. More detailed clinical information can be found in the ‘Control’ Patients section.
Discussion
After in-depth studies of 2 de novo chromosome changes in a girl with severe infantile autism, a 2.6-Mb 2q31.1 deletion and a balanced t(14;21)(q21.1;p11.2) translocation, we have found indications that dysregulation of LRFN5, encoding a synapsis-associated protein, might contribute to the severity of her phenotype with regression of skills since age 2 years. More significantly, this case sheds light on possible epigenetic consequences of balanced translocations.
We have followed the proband from age 1½ years, when her mother came to genetic counselling after discovery of the translocation, to her current age 19. Her development into a severely autistic child was against expectations for a child with a balanced de novo translocation (fig. 1), but the more recent discovery of an apparently unrelated cryptic 2.6-Mb 2q31.1 deletion offered a more plausible explanation for this (fig. 2). This deletion removed (or interrupted) 15 protein-encoding genes: DCAF17, CYBRD1, DYNC1I2, SLC25A12, HAT, MAP1D, DLX1, DLX2, ITGA6, PDK1, RAPGEF4, ZAK, CDCA7, SP3 and OLA1. None of these genes are known to cause mental retardation when haploinsufficient, but RAPGEF4 (Rap guanine nucleotide exchange factor 4) is in theory an interesting candidate due to high expression in the brain and its regulatory role [Kawasaki et al., 1998] (online suppl. fig. 5). However, while RhoGEFs and RhoGTPases have been implicated in mental retardation, also when haploinsufficient [Lybaek et al., 2008, 2009; Nadif Kasri and Van Aelst, 2008], this has, as far as we know, not been shown for RapGEFs. Quite recently patients with overlapping but mostly larger 2q31.1 deletions have been described [Svensson et al., 2007; Mitter et al., manuscript in preparation]. None of these patients were reported to have infantile autism or to have experienced loss of skills, but firm conclusions about such genotype-phenotype relations cannot be drawn, as the patient with the most comparable deletion was only 2 years old at the last examination. However, the strongest argument that our patient's 2q31.1 deletion is not the sole cause of her severe phenotype was her normal development until age 6–7 months. When genomic deletions cause severe mental retardation with autism, signs of poor psychomotor function like lack of eye contact, muscular hypotonia and feeding difficulties are usually apparent from very early age.
We speculated that the t(14;21)(q21.1;p11.2) translocation affected gene expression in the vicinity of the breakpoints by heterochromatin spreading, either from the acrocentric 21p arm into the FBXO33 cluster on derivative chromosome 14, or from the 21p arm/centromere into the LRFN5-containing gene desert on derivative chromosome 21 (fig. 1). After gene expression and ChIP studies in lymphoblasts and skin fibroblasts from the patient and controls, we found that LRFN5 expression was 10-fold reduced in patient fibroblasts compared to LRFN5 expressing control fibroblasts (fig. 3 and online suppl. fig. 3). The complete lack of LRFN5 expression in 3 out of 6 control fibroblasts is so far unexplained, but might relate to REST or miRNA expression (see below) or stochastic promoter changes. Unfortunately, we could not examine patient lymphoblasts for a similar effect because LRFN5 expression was undetectable in this cell type (fig. 3). Lack of expression might be related to the presence of a neuron-restrictive silencer element (NRSE, also called RE1) 323 kb upstream of the LRFN5 promoter [Johnson et al., 2007], i.e. in the middle of the area between the non-coding RNA BX248273 and LRFN5. An RE1-silencing transcription factor (REST) binds to NRSE and represses many neuronal genes in non-neuronal tissues [Jothi et al., 2008].
The reduced LRFN5 expression was correlated with heterochromatinization of the entire LRFN5 locus. Starting exactly at the translocation breakpoint, there was a marked increase in the heterochromatin marker H3K9M3 (figs. 4 and 5). In addition, a patient fibroblast-specific peak of H3K27M3-associated DNA (fig. 4 and online suppl. fig. 4) and a decreased level of H3K4M3-associated DNA (fig. 5) were found at the LRFN5 promoter. It cannot be excluded that both these effects occurred simultaneously, but on 2 different alleles, thereby leading to a full inactivation of the LRFN5 gene in specific tissues. Due to a lack of informative SNPs within the LRFN5 promoter of this patient, we were not able to investigate this possibility. The increase in H3K9M3 level was probably not caused by heterochromatin spreading from the centromere because the centromere 21 with its flanking sequences was intact (fig. 1), and such a centromere is also not known to affect chromatin formation further than ∼0.75 Mb away from the centromeric alpha repeats [Grunau et al., 2006]. Furthermore, in families where 16q11–q13 derived euchromatic material was duplicated and inserted within the major heterochromatin block of chromosome 16 (16qh), there was no evidence for a heterochromatin position effect, i.e. repressed gene expression from the duplicated segment [Barber et al., 2006]. Alternatively, since a centromere is likely to associate with heterochromatin at the nuclear periphery, i.e. in a region of low gene expression [Wiblin et al., 2005], this could have displaced LRFN5 to a region within the nucleus where gene expression was less favoured, and dysregulation could occur, especially since LRFN5 is within the 14q21.2 R-sub-band [Fantes et al., 2008].
LRFN5 is an interesting gene with no known disease associations so far. It belongs to a family of 5 genes that are found only in vertebrates, and that all contain leucine-rich, immunoglobulin- and fibronectin III-like domains [Morimura et al., 2006; Ko and Kim, 2007]. Three LRFN members (1, 2 and 4) have a PSD95 binding domain in the cytoplasmic C-terminal tail [Morimura et al., 2006]. Proteins with such domains are known to influence the postsynaptic density (PSD), a scaffold of proteins important for synaptic differentiation, maintenance and plasticity [Ko et al., 2006]. Mutations in 3 other PSD-associated genes can be rare causes of autism [Jamain et al., 2003; Durand et al., 2007]. LRFN5 has a remarkably isolated location within a 5-Mb region devoid of other protein-encoding genes. Expression is quite brain-specific (online suppl. fig. 5). In mice Lrfn5 gene expression is first turned on after birth [Morimura et al., 2006]. This is the only gene in the LRFN family with 6 exons, the other family members have 2 or 3. The two first of these are non-coding, only contributing to a 5′UTR of nearly 1.2 kb. Such long 5′UTRs are associated with gene regulation at the translational level. The 3′UTR of LRFN5 has several potential micro RNA-family binding sites (see e.g. http://www.targetscan.org/), among which one is a broadly conserved 8-mer binding site for miR-139-5p, a miRNA found to be upregulated in prion-infected mouse brains [Saba et al., 2008]. Taken together, LRFN5's brain-specific expression, genomic isolation, presence of non-coding exons, a long 5′UTR, potential miRNA binding sites in the 3′UTR, and a ncRNA 0.5 Mb upstream, point towards a gene that is tightly regulated, i.e. a gene with a critical function.
We do not yet know if the 10-fold reduction of LRFN5 mRNA expression in patient fibroblasts (fig. 3) is due to random differences in allelic expression levels [Yan et al., 2002], monoallelic expression, like sometimes has been found to cause presumed recessive conditions even though only one allele is lost [Raslova et al., 2004; Mertens et al., 2006], or other forms of allelic interaction leading to inactivation of the other LRFN5 allele. This region of chromosome 14 appears not to be subject to parental-specific imprinting, and neither maternal uniparental disomy 14 [Kotzot, 2004], nor deletion of the LRFN5 locus (in 2 of our control patients) is associated with autism. In the Database of Genomic Variants (http://projects.tcag.ca/) there is only a single CNV affecting the LRFN5 gene; a deletion of the gene's promoter and first 2 exons, detected by a tile-path BAC array [Redon et al., 2006]. So far we have not found any clinical or experimental evidence suggesting that long-range epigenetic silencing of LRFN5 expression should have a phenotypic effect, and it remains possible that the proband's autism with regression of skills has another yet undetected cause, or that her phenotype is at the most severe end of the spectrum for patients with this type of 2q31.1 deletions. However, our finding that a balanced translocation can affect gene expression more than 2 Mb distal to a translocation breakpoint, like the long-range epigenetic silencing that sometimes has been found in tumour cells [Clark, 2007], is of general importance. Such regional effects on gene transcription should be taken into account when considering phenotypic consequences of a de novo balanced translocation. It is also not inconceivable that a familiar translocation might affect gene transcription in one relative but not another, due to random variation in the extent of epigenetic changes around the translocation breakpoint. As a follow-up to our findings, we are now looking for more solid evidence for involvement of LRFN5 in autism, and most importantly, if postzygotic establishment of monoallelic expression might explain why LRFN5 gene deletions are phenotypically neutral.
Supplementary Material
Pictures of the patient taken at ages 9 months and 6 years.
G-banded chromosomes showing the balanced translocation between chromosomes 14 and 21, karyotype 46,XX,t(14;21)(q21.1;p11.2).
Expression analysis of LRFN5 in the patient and three control fibroblasts. LRFN5 expression was readily detected in the control fibroblasts, but was reduced tenfold in the patient fibroblasts. Bar graphs show the level of LRFN5 expression determined by real-time RT-PCR, the scale is logarithmic.
Semi-quantitative PCR for the FBXO33 and LRFN5 promoters on amplified patient fibroblast ChIP material. These data show that the FBXO33 promoter is positive for H3K4M3, but not for H3K27M3. In contrast, the LRFN5 promoter is positive for H3K27M3 and very weakly positive for H3K4M3. These results are in support of the ChIP-on-ChIP data.
Expression pattern of the chromosome 14 genes flanking the t(14;21) translocation breakpoint (FBXO33 and LRFN5), and selected genes from the 2q31 microdeletion (SLC25A12, DLX1 and RAPGEF4). Expression data was taken from the publicly available GNF dataset (http://biogps.gnf.org).
Supplemental Table
Acknowledgements
We would like to thank Niels Tommerup who helped us get EBV-transformed lymphoblasts from the patient, and Saskia van der Velde-Visser and Christel Beumer for their help with cell culturing. We also thank Christian Gilissen and Ad Geurts van Kessel for advice and support. Finally, we are most grateful to the patient's mother, whose enthusiastic collaboration made our work possible.
References
- Baptista J, Mercer C, Prigmore E, Gribble SM, Carter NP, et al. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am J Hum Genet. 2008;82:927–936. doi: 10.1016/j.ajhg.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber JC, Zhang S, Friend N, Collins AL, Maloney VK, et al. Duplications of proximal 16q flanked by heterochromatin are not euchromatic variants and show no evidence of heterochromatic position effect. Cytogenet Genome Res. 2006;114:351–358. doi: 10.1159/000094225. [DOI] [PubMed] [Google Scholar]
- Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum Mol Genet. 2007;16(Spec No 1):R88–R95. doi: 10.1093/hmg/ddm051. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- de Bruijn DR, Kater-Baats E, Eleveld M, Merkx G, Geurts Van Kessel A. Mapping and characterization of the mouse and human SS18 genes, two human SS18-like genes and a mouse Ss18 pseudogene. Cytogenet Cell Genet. 2001;92:310–319. doi: 10.1159/000056920. [DOI] [PubMed] [Google Scholar]
- de Bruijn DR, Allander SV, van Dijk AH, Willemse MP, Thijssen J, et al. The synovial-sarcoma-associated SS18-SSX2 fusion protein induces epigenetic gene (de)regulation. Cancer Res. 2006;66:9474–9482. doi: 10.1158/0008-5472.CAN-05-3726. [DOI] [PubMed] [Google Scholar]
- De Gregori M, Ciccone R, Magini P, Pramparo T, Gimelli S, et al. Cryptic deletions are a common finding in ‘balanced’ reciprocal and complex chromosome rearrangements: a study of 59 patients. J Med Genet. 2007;44:750–762. doi: 10.1136/jmg.2007.052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K, Vanhole C, Matthijs G, de Zegher F. Deletion of thyroid transcription factor-1 gene in an infant with neonatal thyroid dysfunction and respiratory failure. N Engl J Med. 1998;338:1317–1318. doi: 10.1056/NEJM199804303381817. [DOI] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes JA, Boland E, Ramsay J, Donnai D, Splitt M, et al. FISH mapping of de novo apparently balanced chromosome rearrangements identifies characteristics associated with phenotypic abnormality. Am J Hum Genet. 2008;82:916–926. doi: 10.1016/j.ajhg.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RJM, Sutherland GR. Chromosome Abnormalities and Genetic Couseling. 3rd ed. New York: Oxford University Press; 2004. [Google Scholar]
- Grunau C, Buard J, Brun ME, De Sario A. Mapping of the juxtacentromeric heterochromatin-euchromatin frontier of human chromosome 21. Genome Res. 2006;16:1198–1207. doi: 10.1101/gr.5440306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H, Springett GM, Toki S, Canales JJ, Harlan P, et al. A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc Natl Acad Sci USA. 1998;95:13278–13283. doi: 10.1073/pnas.95.22.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Kim E. Leucine-rich repeat proteins of synapses. J Neurosci Res. 2007;85:2824–2832. doi: 10.1002/jnr.21306. [DOI] [PubMed] [Google Scholar]
- Ko J, Kim S, Chung HS, Kim K, Han K, et al. SALM synaptic cell adhesion-like molecules regulate the differentiation of excitatory synapses. Neuron. 2006;50:233–245. doi: 10.1016/j.neuron.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Kotzot D. Maternal uniparental disomy 14 dissection of the phenotype with respect to rare autosomal recessively inherited traits, trisomy mosaicism, and genomic imprinting. Ann Genet. 2004;47:251–260. doi: 10.1016/j.anngen.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krude H, Schutz B, Biebermann H, von Moers A, Schnabel D, et al. Choreoathetosis, hypothyroidism, and pulmonary alterations due to human NKX2-1 haploinsufficiency. J Clin Invest. 2002;109:475–480. doi: 10.1172/JCI14341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AW, Liao GC, Rocchi M, Choo KH. Extreme reduction of chromosome-specific alpha-satellite array is unusually common in human chromosome 21. Genome Res. 1999;9:895–908. doi: 10.1101/gr.9.10.895. [DOI] [PubMed] [Google Scholar]
- Lybaek H, Oyen N, Fauske L, Houge G. A 2.1 Mb deletion adjacent but distal to a 14q21q23 paracentric inversion in a family with spherocytosis and severe learning difficulties. Clin Genet. 2008;74:553–559. doi: 10.1111/j.1399-0004.2008.01072.x. [DOI] [PubMed] [Google Scholar]
- Lybaek H, Orstavik KH, Prescott T, Hovland R, Breilid H, et al. An 8.9 Mb 19p13 duplication associated with precocious puberty and a sporadic 3.9 Mb 2q23.3q24.1 deletion containing NR4A2 in mentally retarded members of a family with an intrachromosomal 19p-into-19q between-arm insertion. Eur J Hum Genet. 2009;17:904–910. doi: 10.1038/ejhg.2008.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens D, Wolf S, Tschuch C, Mund C, Kienle D, et al. Allelic silencing at the tumor-suppressor locus 13q14.3 suggests an epigenetic tumor-suppressor mechanism. Proc Natl Acad Sci USA. 2006;103:7741–7746. doi: 10.1073/pnas.0600494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura N, Inoue T, Katayama K, Aruga J. Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene. 2006;380:72–83. doi: 10.1016/j.gene.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Nadif Kasri N, Van Aelst L. Rho-linked genes and neurological disorders. Pflugers Arch. 2008;455:787–797. doi: 10.1007/s00424-007-0385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannya Y, Sanada M, Nakazaki K, Hosoya N, Wang L, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Raslova H, Komura E, Le Couedic JP, Larbret F, Debili N, et al. FLI1 monoallelic expression combined with its hemizygous loss underlies Paris-Trousseau/Jacobsen thrombopenia. J Clin Invest. 2004;114:77–84. doi: 10.1172/JCI21197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluth-Bolard C, Delobel B, Sanlaville D, Boute O, Cuisset JM, et al. Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: array CGH study of 47 unrelated cases. Eur J Med Genet. 2009;52:291–296. doi: 10.1016/j.ejmg.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Sismani C, Kitsiou-Tzeli S, Ioannides M, Christodoulou C, Anastasiadou V, et al. Cryptic genomic imbalances in patients with de novo or familial apparently balanced translocations and abnormal phenotype. Mol Cytogenet. 2008;1:15. doi: 10.1186/1755-8166-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson AM, Curry CJ, South ST, Whitby H, Maxwell TM, et al. Detection of a de novo interstitial 2q microdeletion by CGH microarray analysis in a patient with limb malformations, microcephaly and mental retardation. Am J Med Genet A. 2007;143A:1348–1353. doi: 10.1002/ajmg.a.31775. [DOI] [PubMed] [Google Scholar]
- Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet. 1991;49:995–1013. [PMC free article] [PubMed] [Google Scholar]
- Wiblin AE, Cui W, Clark AJ, Bickmore WA. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci. 2005;118:3861–3868. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pictures of the patient taken at ages 9 months and 6 years.
G-banded chromosomes showing the balanced translocation between chromosomes 14 and 21, karyotype 46,XX,t(14;21)(q21.1;p11.2).
Expression analysis of LRFN5 in the patient and three control fibroblasts. LRFN5 expression was readily detected in the control fibroblasts, but was reduced tenfold in the patient fibroblasts. Bar graphs show the level of LRFN5 expression determined by real-time RT-PCR, the scale is logarithmic.
Semi-quantitative PCR for the FBXO33 and LRFN5 promoters on amplified patient fibroblast ChIP material. These data show that the FBXO33 promoter is positive for H3K4M3, but not for H3K27M3. In contrast, the LRFN5 promoter is positive for H3K27M3 and very weakly positive for H3K4M3. These results are in support of the ChIP-on-ChIP data.
Expression pattern of the chromosome 14 genes flanking the t(14;21) translocation breakpoint (FBXO33 and LRFN5), and selected genes from the 2q31 microdeletion (SLC25A12, DLX1 and RAPGEF4). Expression data was taken from the publicly available GNF dataset (http://biogps.gnf.org).
Supplemental Table