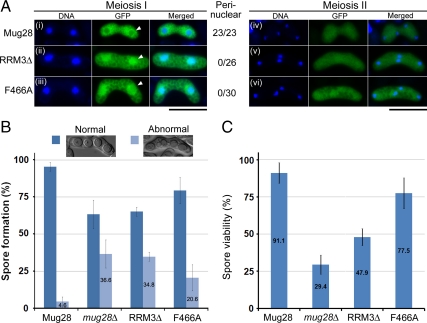
Figure 9.
Functional analysis of the Mug28F466A mutant. (A) Subcellular localization of GFP-tagged Mug28 protein (green) during meiosis I and meiosis II in mug28Δ (AS99) cells: Mug28-GFP (AS164; i and iv), Mug28RRM3Δ-GFP (AS176; ii and v), and Mug28F466A-GFP (NP252; iii and vi). Merged images with DNA (blue) are shown in the rightmost panels. Arrowheads indicate normal (i) or abnormal localization of Mug28RRM3Δ-GFP (ii) and Mug28F466A-GFP (iii) to the perinucleus during meiosis I. Numbers represent the proportion of cells displaying perinuclear GFP signals per examined cell. Bar, 10 μm. (B) Frequency of normal and abnormal spore formation in WT (Mug28) and mutants (mug28Δ, RRM3Δ, and F466A). Bar graphs show the averages of three independent experiments with SEs. At least 200 spores were measured for each strain. (C) Spore viability of WT (Mug28) and mutants (mug28Δ, RRM3Δ, and F466A), measured by random spore analysis. Bar graphs show the averages of three independent experiments with SEs. At least 200 spores were measured for each strain.