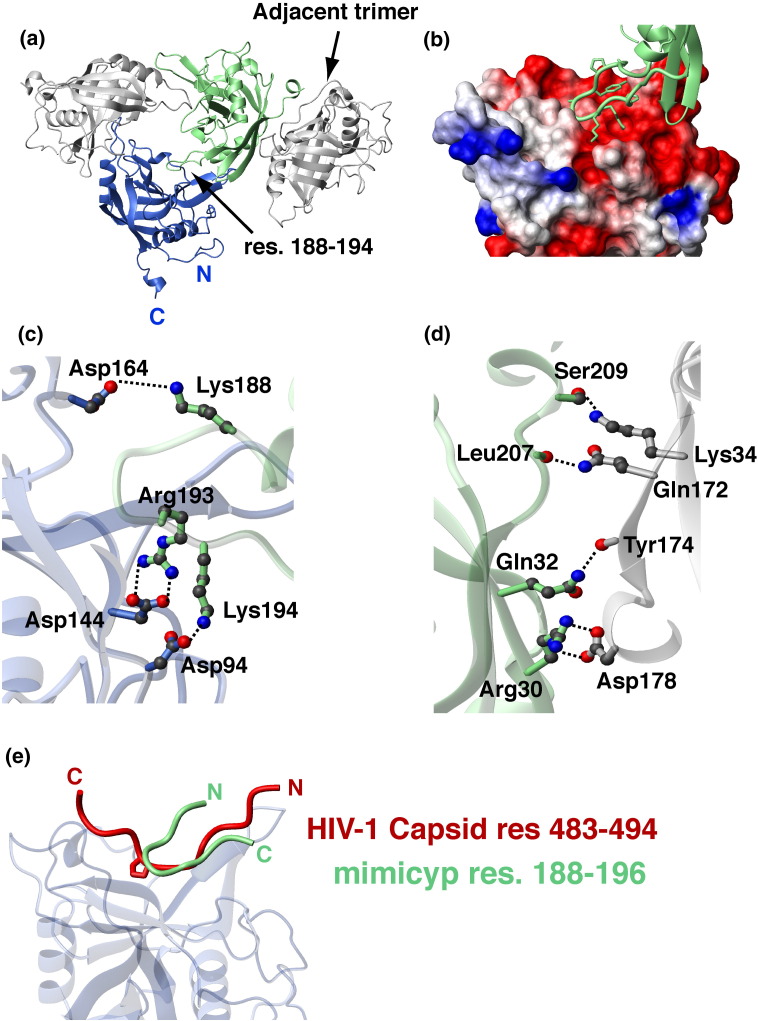
Figure 2.
Crystallographic interactions of mimicyp and comparison to hCypA interactions with the HIV-1 capsid. (a) MimiCyp crystallizes with four trimers within each unit cell. Each monomer within the trimer interacts via a basic loop insertion, residues 188–194 (KPYAGRK) into the putative active site of a neighboring monomer. Residues on the opposite side of the monomer from the putative active site, including residues within the C-terminal helix, mediate trimer–trimer contacts. (b) Close-up of the basic loop insertion into the acidic putative active site that forms each trimer. The basic loop is shown in green with heavy atoms of side-chains included and the active site is shown as a surface representation with basic residues shown in blue, acidic residues in red, and uncharged or hydrophobic residues in white. (c) Intermolecular electrostatic side-chain pairs that stabilize the trimer are formed between Lys188, Arg193, Lys194 from the basic loop and Asp164, Asp144 and Asp94 of the putative acidic active site, respectively. (d) Intermolecular electrostatic pairs that stabilize trimer–trimer formation include side-chain interactions of Asp178–Arg30 and Ser209–Lys34, and intermolecular hydrogen bonds formed between backbone carbonyl oxygen atoms of Tyr174 and Leu207 with the side-chains of Gln32 and Gln172, respectively. (e) Overlay of mimicyp intermolecular interactions with the hCypA/HIV-1 capsid complex (PDB accession number 1AK4) exhibits both similarities and differences. The mimicyp residues 188–194 are shown that comprise a type II β-turn, Tyr190–Arg193, and the HIV-1 capsid residues 483–494 form a relatively extended structure. These loops bind to each cyclophilin in opposing directions, yet approximately half of the residues within each, i.e. comprising the HIV-1 capsid loop and the neighboring mimicyp inserted loop, are nearly superimposed. The same secondary structural elements of mimicyp and hCypA shown in Figure 1 were used to align the two complexes.