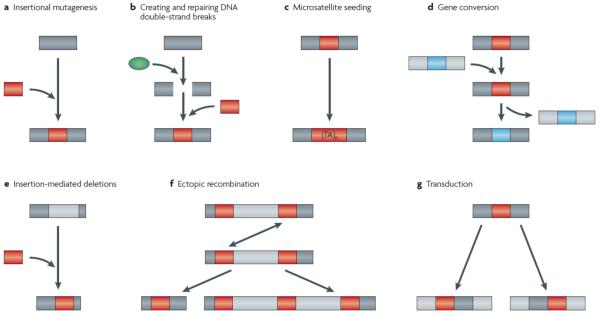
Figure 2. Impact of retrotransposons on human genome structure.
a | Typical insertion of an L1, Alu or SVA retrotransposon (red box) at a new genomic site (blue area). If the new genomic site is a genic region, the retrotransposon may cause insertional mutagenesis. b | The protein products of an L1 element (red circle) may create DNA double-strand breaks (broken blue area). Alternatively, an existing double-strand break may be repaired via non-classical endonuclease-independent insertion of a retrotransposon (red box). c | Microsatellites (e.g. (TA)n) may arise from the homopolymeric tracts endogenous to retrotransposons. d | Gene conversion may alter the sequence compositions of homologous retrotransposon copies (red and green boxes). e | Insertion of retrotransposons (red box) is sometimes associated with concomittant deletion of target genomic sequences (light blue box). f | Ectopic recombination (double arrowheaded line) between non-allelic homologous retrotransposons (red boxes) may result in genomic reaarangements such as deletions (left) or duplications (right) of intervening genomic sequences. g | 3′ and 5′ transduction can result in the co-retrotransposition of downstream or upstream flanking genomic sequence, respectively, along with a retrotransposon (left and right, respectively).