Figure 1. Conserved non-coding sequences and chromatin modifications at the Foxp3 locus.
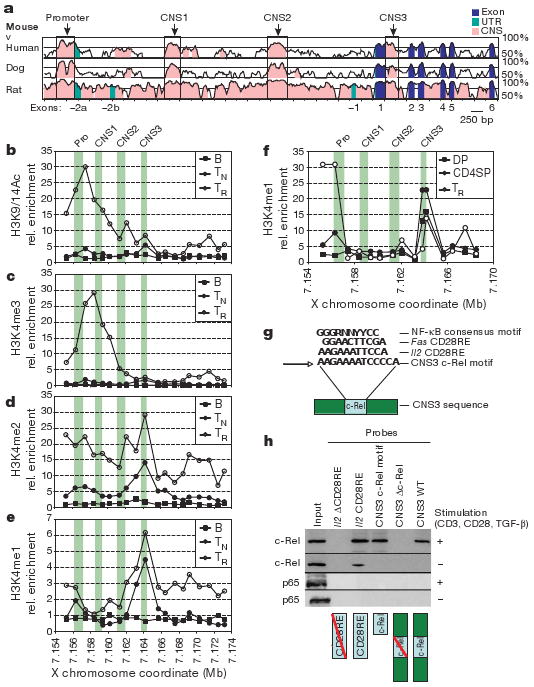
a, Comparison of mouse Foxp3 genomic sequence to human, dog and rat. b–e, Map of permissive chromatin modifications at the Foxp3 locus by ChIP–qPCR with primers spaced at ∼1-kb intervals for B220+ B cells (B), CD4+CD25− TN cells, and CD4+CD25+ Treg (TR) cells. Relative (rel.) enrichment data are shown for H3K9/14Ac (b), H3K4me3 (c), H3K4me2 (d) and H3K4me1 (e). f, H3K4me1 ChIP–qPCR as in e, for CD4+CD8+ (double positive, DP) and CD4+CD8− (CD4 single positive, CD4SP) thymocytes, and Treg cells. In b–f, green bars denote the promoter (Pro) and CNS1–3. g, NF-κB consensus motif in CNS3 (core motif in bold) and homologous CD28RE motifs from Il2 and Fas. N, any base; R, purine; Y, pyrimidine. h, Binding of NF-κB family member c-Rel but not p65 to the CD28RE-like element at CNS3. Nuclear lysates from unstimulated or stimulated (1 μg ml−1 CD3 and CD28 antibodies, 2 ng ml−1 TGF-β) TN cells were incubated with biotinylated double-stranded (ds)DNA probes containing the full-length CNS3 sequence (CNS3 WT), the full-length CNS3 sequence with the mutated core c-Rel motif (CNS3 Δc-Rel), the core CNS3 c-Rel motif, Il2 CD28RE (positive control), and Il2 ΔCD28RE (negative control). dsDNA probes and bound protein were precipitated by streptavidin beads and subjected to c-Rel and p65 western blot analysis.
