Figure 1.
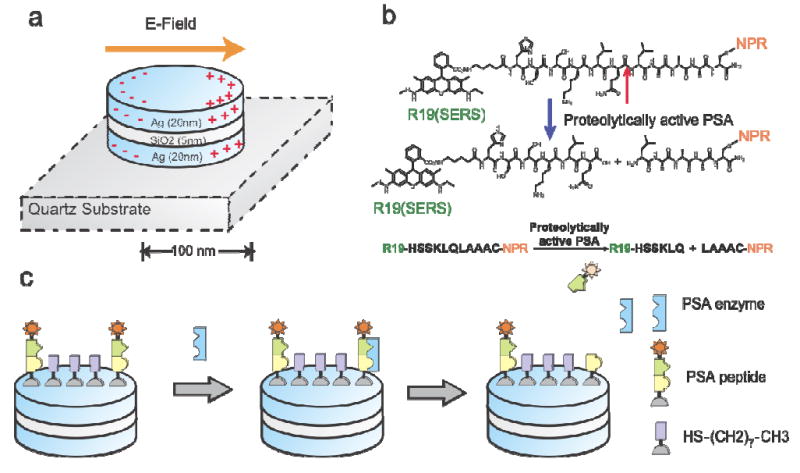
Schematic illustration of the working principle of detecting PSA protease activity using peptide-conjugated NPR SERS nanosensors. (a) NPRs exhibit a tunable plasmon resonance and highly enhanced local electromagnetic field through coupled plasmonic resonance. NPRs with a short axis of 150 nm and long axis of 200 nm were made of multi-stacks of silver and SiO2 layers with thicknesses of 25 nm and 5 nm, respectively. (b) The molecular structure of the biomarker that consists of Raman dye R19, PSA specific peptide sequence HSSKLQLAAAC, and cysteine. The peptide can be cleaved by PSA enzyme between HSSKLQ and LAAAC. (c) The detection scheme of NPR functionalized with peptide sequence HSSKLQLAAAC and the Raman dye R19 (star). The presence of the PSA enzyme will cleave the peptide sequence. After cleavage, the diffusion of the R19 (star) away from the surface will be monitored by the loss of the SERS signatures of the R19 moiety. Packing molecule octanethiol (HS-(CH2)2-CH3) was used to reduce the packing density of the reporting peptide on NPR surface, and thus, allows PSA enzyme to access the reporting peptide.
