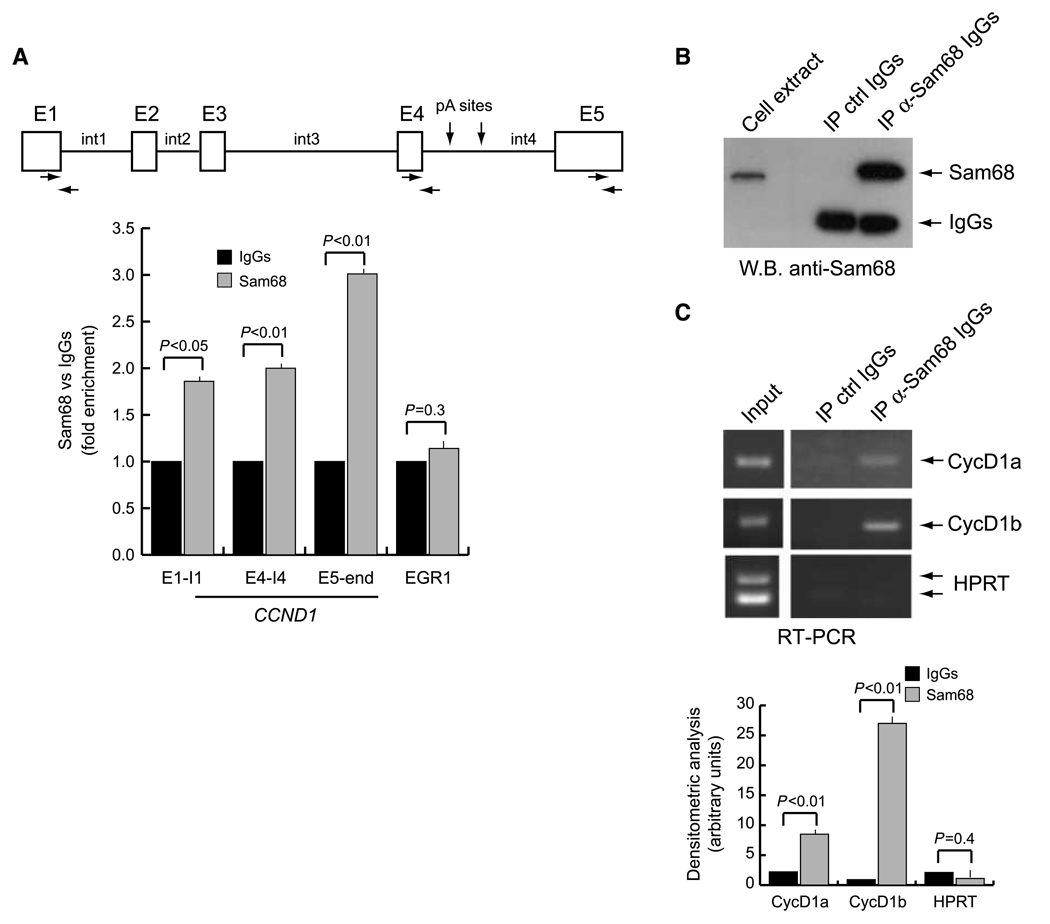
Figure 1.
Sam68 associates with CCND1 gene and with cyclin D1 mRNA in LNCaP cells. A, scheme of the CCND1 gene. Boxes identify the five exons (E1–E5), whereas the lines identify the four introns (int1–int4). Horizontal arrows, position of the primers used for the PCR of the immunoprecipitated chromatin; vertical arrows, position of the polyadenylation sites in intron 4 (28). Cross-linked chromatin derived from LNCaP cells was immunoprecipitated with anti-Sam68 antibody or control IgGs. The immunoprecipitated chromatin was analyzed by quantitative real-time PCR using the indicated primers. Early growth response 1 (EGR1) primers were used as internal control for a gene not regulated by Sam68. Results are shown as fold enrichment in the anti-Sam68 immunoprecipitates versus the control IgGs. B and C, coimmunoprecipitation of endogenous Sam68 with cyclin D1 mRNA. LNCaP cell extracts were immunoprecipitated with anti–Sam68-specific antibody; IgGs were used as mock reaction; coprecipitated RNAs were analyzed by RT-PCR with primers specific for cyclin D1a (CycD1a), cyclin D1b (CycD1b), and hypoxanthine phosphoribosyltransferase (HPRT) used as negative control (C). An aliquot of the immunoprecipitates was saved for Western blot analysis of Sam68 (B). A and C, columns, mean of three independent experiments; bars, SD. P values of Student's t test are reported.