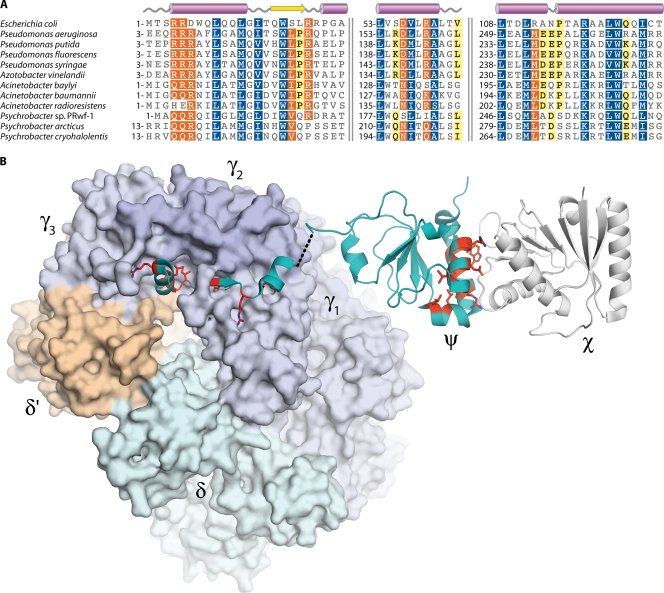
FIG. 2.
Sequence conservation within the ψ subunit of DNA polymerase III. (A) Multiple alignment of ψ amino acid sequences. Numbers indicate the sequence position of the first residue in each alignment block; pairs of vertical lines denote discontinuities in the sequences. White characters with blue shading indicate positions with homology across all sequences. White characters with orange shading indicate residues conserved in 80% of sequences, while black characters with yellow shading indicate 60% conservation. The sequence alignment was produced using the MUSCLE algorithm (72) within the Geneious software package (Biomatters). (B) Crystal structures of the E. coli γ3δδ′ψ and χψ complexes. Residues conserved in the A. baylyi ψ subunit are colored red and have side chains shown. (Produced with PyMOL [64] using data from Protein Data Bank entries 3GLI [231] and 1EM8 [96].)