Abstract
Enterotoxigenic Escherichia coli (ETEC) is a common pathogen worldwide causing infectious diarrhea, especially traveler's diarrhea. Traditional physiological assays, immunoassays, and PCR-based methods for the detection of ETEC target the heat-labile enterotoxin and/or the heat-stable enterotoxin. Separate serotyping methods using antisera are required to determine the ETEC serogroup. In this study, we developed a DNA microarray that can simultaneously detect enterotoxin genes and the 19 most common O serogroup genes in ETEC strains. The specificity and reproducibility of this approach were verified by hybridization to 223 strains: 50 target reference or clinical strains and 173 other strains, including those belonging to other E. coli O serogroups and closely related species. The sensitivity of detection was determined to be 50 ng of genomic DNA or 108 CFU per ml of organisms in pure culture. The random PCR strategy used in this study with minimal bias provides an effective alternative to multiplex PCR for the detection of pathogens using DNA microarrays. The assay holds promise for applications in the clinical diagnosis and epidemiological surveillance of pathogenic microorganisms.
Enterotoxigenic Escherichia coli (ETEC) is the leading bacterial cause of infectious diarrhea in the developing world, causing infantile or cholera-like disease in all age groups (2). It is among the major etiologic agents, leading to an estimated 1.5 million deaths per year worldwide (13, 14). ETEC is also a major cause of traveler's diarrhea (3, 8, 11) and the most common pathogen among the six recognized diarrheagenic categories of E. coli, especially in the developing world (18). ETEC strains produce one or both of the following two enterotoxins: heat-labile enterotoxin (LT) and heat-stable enterotoxin (ST). Two classes of STs—STa and STb—and two variants of STa—STp (initially discovered in isolates from pigs) and STh (initially discovered in isolates from humans)—have been described. The elt, estA, and estB genes encode the enterotoxins LT, STa, and STb, respectively (6, 23, 26).
The O antigen comprises the outermost domain of the lipopolysaccharide molecule and is attached to the core oligosaccharide on the surfaces of Gram-negative bacteria (20). O antigens are among the most variable cellular constituents, imparting antigenic specificity. The composition of the O chain differs from strain to strain; more than 180 O-antigen structures are produced by different E. coli strains (25). The most common O serogroups reported in ETEC are O6, O8, O11, O15, O25, O27, O78, O85, O114, O115, O126, O128, O139, O148, O149, O159, O166, O167, and O173 (5, 18, 19, 31).
Detection of ETEC has long relied on detection of the enterotoxins LT and/or ST by physiological assays and immunoassays, and serotyping has depended on assays using O-serogroup-specific antisera. These traditional approaches are slow and labor-intensive, and assays using antisera can be impeded by cross-reactivity. PCR assays, which are more rapid, sensitive, and specific, have also been widely used for ETEC diagnosis (15, 24). However, molecular methods for the serotyping of ETEC have not been developed.
Molecular detection and typing by PCR and microarray techniques have many advantages over traditional methods. DNA microarrays provide an efficient approach for the parallel detection and analysis of a large number of pathogenic microorganisms. This technique has been applied to the detection of pathogens from all kinds of biological samples, including water, food, and soil (4, 7, 12, 17, 21).
In this study, we developed a DNA microarray for the detection and typing of ETEC. The genes encoding the enterotoxins LT and ST were used for the detection of ETEC, and the serogroup-specific genes wzx and/or wzy were used for the typing of the 19 most common ETEC O serogroups. The microarray was examined for its specificity and sensitivity, and the findings of this study indicate that it is highly sensitive and reproducible.
MATERIALS AND METHODS
Bacterial strains.
The 223 strains used in this study are listed in Table 1 . They include 28 reference strains and 22 clinical strains of the 19 targeted E. coli O serogroups (O6, O8, O11, O15, O25, O27, O78, O85, O114, O115, O126, O128, O139, O148, O149, O159, O166, O167, and O173) and 150 reference strains of other E. coli O serogroups. Also included in this study were 13 reference strains of different Shigella O serogroups and 10 reference strains of different Salmonella O serogroups. All strains were grown overnight in Luria-Bertani medium at 37°C with shaking.
TABLE 1.
Strains used in this study
| Strain no. | Sourcea | Serogroup or serotypeb | Virulence factor(s) |
|---|---|---|---|
| E. coli strains with targeted O serogroups | |||
| Reference strains | |||
| G1062 | a | O6 | None |
| G1640 | a | O6 | None |
| G1654 | a | O6 | None |
| G1602 | a | O8 | None |
| G1650 | a | O11 | None |
| G1657 | a | O11 | None |
| G1130 | b | O11 | None |
| G1201 | a | O15 | None |
| G1249 | a | O25 | None |
| G1111 | b | O25 | None |
| G1286 | a | O27 | None |
| G1235 | a | O78 | None |
| G1189 | a | O85 | None |
| G1160 | b | O85 | None |
| G1088 | a | O114 | None |
| G1695 | a | O115 | None |
| G1679 | a | O126 | None |
| G1095 | a | O128 | None |
| G1208 | a | O139 | None |
| G1122 | b | O139 | None |
| G1258 | a | O148 | None |
| G1127 | b | O148 | STh |
| G1392 | a | O148 | None |
| G1061 | a | O149 | None |
| G1108 | a | O159 | None |
| G1216 | a | O166 | None |
| G1185 | a | O167 | STh |
| G1093 | a | O173 | None |
| Clinical isolates | |||
| 151/05/G2493 | c | O8 | LT, STb |
| CB08768/G2521 | c | O8 | STp |
| CB08810/G2533 | c | O8 | STp |
| Bi 623-42/G1306 | c | O11 | None |
| F7902-41/G1383 | c | O15 | None |
| 2P9/G1384 | c | O15 | None |
| CA017(19)/G1385 | c | O15 | None |
| RL84/98/G2579 | c | O78 | None |
| RL453/98/G2580 | c | O78 | None |
| RL415/98/G2581 | c | O78 | None |
| RL468/98/G2582 | c | O78 | None |
| C275-53/G1379 | c | O114 | STh |
| 339-54/G1361 | c | O114 | LT |
| C1003-63/G1336 | c | O114 | STh |
| 3075/69/G1341 | c | O114 | None |
| 340-54/G1362 | c | O114 | LT |
| IP831/G1325 | c | O114 | LT |
| 150/05/G2492 | c | O149 | LT, STb |
| 205/05/G2503 | c | O149 | LT, STp, STb |
| 494/99/G2523 | c | O149 | LT, STp, STb |
| 425/98/G2524 | c | O149 | LT, STb |
| 376/98/G2525 | c | O149 | LT, STp, STb |
| E. coli reference strains with nontargeted O serogroups | |||
| G1673 | a | O1 | |
| G1674 | a | O2 | |
| G1206 | a | O3 | |
| G1633 | a | O4 | |
| G1675 | a | O5 | |
| G1676 | a | O7 | |
| G1677 | a | O9 | |
| G1055 | a | O10 | |
| G1280 | a | O12 | |
| G1237 | a | O13 | |
| G1678 | a | O14 | |
| G1680 | a | O16 | |
| G1298 | a | O17 | |
| G1299 | a | O18ac | |
| G1202 | a | O19ab | |
| G1501 | a | O20 | |
| G1210 | a | O21 | |
| G1681 | a | O22 | |
| G1199 | a | O23 | |
| G1204 | a | O24 | |
| G1682 | a | O26 | |
| G1683 | a | O28 | |
| G1188 | a | O29 | |
| G1684 | a | O30 | |
| G1264 | a | O32 | |
| G1195 | a | O33 | |
| G1063 | a | O34 | |
| G1211 | a | O35 | |
| G1064 | a | O36 | |
| G1241 | a | O37 | |
| G1685 | a | O38 | |
| G1056 | a | O39 | |
| G1234 | a | O40 | |
| G1289 | a | O41 | |
| G1065 | a | O42 | |
| G1247 | a | O43 | |
| G1291 | a | O44 | |
| G1686 | a | O45 | |
| G1687 | a | O46 | |
| G1692 | a | O91 | |
| G1079 | a | O92 | |
| G1080 | a | O95 | |
| G1081 | a | O96 | |
| G1082 | a | O97 | |
| G1083 | a | O98 | |
| G1251 | a | O99 | |
| G1058 | a | O100 | |
| G1502 | a | O101 | |
| G1240 | a | O102 | |
| G1693 | a | O103 | |
| G1629 | a | O104 | |
| G1084 | a | O105 | |
| G1255 | a | O106 | |
| G1085 | a | O107 | |
| G1198 | a | O108 | |
| G1259 | a | O109 | |
| G1086 | a | O110 | |
| G1087 | a | O111 | |
| G1295 | a | O112ab | |
| G1694 | a | O113 | |
| G1261 | a | O116 | |
| G1089 | a | O117 | |
| G1696 | a | O118 | |
| G1059 | a | O119 | |
| G1293 | a | O120 | |
| G1060 | a | O121 | |
| G1697 | a | O123 | |
| G1053 | b | O124 | |
| G1209 | a | O125 | |
| G1094 | a | O127 | |
| G1096 | a | O129 | |
| G1203 | a | O130 | |
| G1193 | a | O131 | |
| G1297 | a | O132 | |
| G1272 | a | O133 | |
| G1626 | a | O48 | |
| G1277 | a | O49 | |
| G1688 | a | O50 | |
| G1218 | a | O51 | |
| G1066 | a | O52 | |
| G1067 | a | O53 | |
| G1245 | a | O54 | |
| G1284 | a | O55 | |
| G1068 | a | O56 | |
| G1069 | a | O57 | |
| G1248 | a | O58 | |
| G1070 | a | O59 | |
| G1627 | a | O60 | |
| G1071 | a | O61 | |
| G1290 | a | O62 | |
| G1072 | a | O63 | |
| G1073 | a | O64 | |
| G1279 | a | O65 | |
| G1074 | a | O66 | |
| G1215 | a | O68 | |
| G1628 | a | O69 | |
| G1635 | a | O70 | |
| G1194 | a | O71 | |
| G1057 | a | O73 | |
| G1231 | a | O74 | |
| G1689 | a | O75 | |
| G1220 | a | O76 | |
| G1075 | a | O77 | |
| G1690 | a | O79 | |
| G1076 | a | O80 | |
| G1217 | a | O81 | |
| G1196 | a | O82 | |
| G1691 | a | O83 | |
| G1205 | a | O84 | |
| G1275 | a | O86 | |
| G1260 | a | O87 | |
| G1262 | a | O88 | |
| G1077 | a | O89 | |
| G1078 | a | O90 | |
| G1253 | a | O134 | |
| G1239 | a | O135 | |
| G1265 | a | O136 | |
| G1276 | a | O137 | |
| G1242 | a | O138 | |
| G1097 | a | O140 | |
| G1230 | a | O141 | |
| G1098 | a | O142 | |
| G1288 | a | O143 | |
| G1099 | a | O144 | |
| G1100 | a | O145 | |
| G1101 | a | O146 | |
| G1229 | a | O147 | |
| G1102 | a | O150 | |
| G1103 | a | O151 | |
| G1104 | a | O152 | |
| G1105 | a | O153 | |
| G1273 | a | O154 | |
| G1106 | a | O155 | |
| G1197 | a | O156 | |
| G1704 | a | O157 | |
| G1107 | a | O158 | |
| G1109 | a | O160 | |
| G1254 | a | O161 | |
| G1283 | a | O162 | |
| G1244 | a | O163 | |
| G1225 | a | O164 | |
| G1698 | a | O165 | |
| G1090 | a | O168 | |
| G1278 | a | O169 | |
| G1091 | a | O170 | |
| G1270 | a | O171 | |
| G1092 | a | O172 | |
| G1609 | c | O174 | |
| G1606 | c | O177 | |
| G2258 | c | O180 | |
| Strains of other bacterial species (n = 23) | |||
| Shigella species | |||
| Shigella flexneri G1661 | a | Type 1a | |
| S. flexneri G1663 | a | Type 2a | |
| S. flexneri G1665 | a | Type 3a | |
| S. flexneri G1668 | a | Type 4a | |
| S. flexneri G1669 | a | Type 4b | |
| S. dysenteriae G1252 | a | Type 2 | |
| S. dysenteriae G1281 | a | Type 3 | |
| S. dysenteriae G1213 | a | Type 5 | |
| Shigella boydii G1296 | a | Type 4 | |
| S. boydii G1236 | a | Type 9 | |
| S. dysenteriae G1018 | d | Type 1 | |
| S. dysenteriae G2547 | b | Type 1 | |
| S. dysenteriae G2548 | b | Type 1 | |
| Salmonella enterica | |||
| G1440 | a | Serotype Dakar | |
| G1441 | a | Serotype Utrecht | |
| G1450 | a | Serotype Anatum | |
| G1459 | a | Serotype Hvittingfoss | |
| G1460 | a | Serotype Jangwani | |
| G1462 | a | Serotype II 47:b:1,5 | |
| G1465 | a | Serotype Urbana | |
| G1467 | a | Serotype Niarembe | |
| G1481 | a | Serogroup D1 | |
| G1805 | a | Serotype Marseille | |
Lowercase letters represent sources as follows: a, Institute of Medical and Veterinary Science, Adelaide, Australia; b, National Center for Medical Culture Collection, China; c, Federal Institute for Risk Assessment (BfR), Berlin, Germany; d, National Institute for Communicable Disease Control and Prevention, Beijing, China.
Serogroups are given for E. coli strains.
Genomic DNA extraction.
Bacterial genomic DNA was extracted using the TIANamp Bacteria DNA kit (Tiangen, Beijing, China).
DNA amplification and labeling using the random PCR method.
The first step was performed as follows: a reaction mixture containing 1 μl of 100-ng/μl genomic DNA, 1 μl of 100 μM primer A (5′-GTTTCCCAGTCACGATCNNNNNNNNN-3′) (22), and 8 μl of Milli-Q water was first incubated at 95°C for 5 min and then cooled to 4°C for 2 to 5 min. The reaction mixture was then made up to 30 μl by the addition of 4 μl of 10× PCR buffer (500 mM KCl, 100 mM Tris-HCl [pH 8.3], 5 mM MgCl2), 1 μl of 10 mM deoxynucleoside triphosphates (dNTPs), 2.5 U of Taq DNA polymerase, and an appropriate volume of Milli-Q water. The mixture was kept at 4°C for 10 s. Then the primer was allowed to anneal by slowly increasing the temperature from 4 to 72°C over a 10-min period, followed by an extension step at 72°C for 1 min. The steps described above were repeated once, and the PCR products were then purified using the Montage centrifugal filter device kit (Millipore Corporation, MA).
The second step was performed with a 20-μl reaction mixture consisting of 6 μl of the purified PCR product from the first step, 1× PCR buffer, 0.2 mM dNTPs, 0.1 U of Taq DNA polymerase, 2 μM primer B (5′-GTTTCCCAGTCACGATC-3′), and Milli-Q water (22). The reaction parameters were as follows: 95°C for 5 min; 35 cycles of 95°C for 30 s, 45°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final extension at 72°C for 5 min. A 2-μl aliquot of the resulting PCR product was run on an agarose gel to check if the amplified DNA appeared as a 250- to 1,000-bp smear. This DNA was then used as the template for labeling.
The third step was performed with a 40-μl reaction mixture consisting of 5 μl of the PCR product from the first step, 1× PCR buffer, 0.25 mM dNTPs, 2.5 μM primer B, 0.125 U Taq DNA polymerase, and 0.3125 nM Cy3-dUTP. The reaction parameters were the same as those described for the second step.
Oligonucleotide probe design.
For each serogroup, two probes were designed for OligoArray, version 2.0, based on sequences in the GenBank database and an in-house database consisting of all 34 of the O-antigen gene clusters of Shigella and 175 O-antigen gene clusters of E. coli. For each virulence gene (elt, estAp, estAh, and estB), two probes were designed. Two probes based on the rfpB gene of Shigella dysenteriae type 1 were designed to differentiate E. coli O148 from S. dysenteriae type 1. One probe based on bacterial 16S rRNA genes was designed as a positive control. A probe containing 40 poly(T) oligonucleotides was used as a negative control. A probe labeled with Cy3 at the 3′ end was used as the positional reference and printing control. Each probe was 5′ amino modified, and 10 poly(T) oligonucleotides were added [for the probe based on 16S rRNA genes, 15 poly(T) oligonucleotides were added]. All of the oligonucleotide probes used are listed in Table 2.
TABLE 2.
Oligonucleotide probes used in this study
| Probe | Targeted gene (serogroup) | Source/GenBank accession no. | Tm (°C)a | Sequence (5′-3′) |
|---|---|---|---|---|
| OA-2443 | wzx (O6) | NC_004431 | 79.86 | CCATGTTGTTCATCTTAAACCTAATGAATGCATTGTGGAA |
| OA-2444 | wzy (O6) | NC_004431 | 79.62 | TCGTAGTGAAGCTATAACGTTTCTTTTAACGGTTACATGT |
| OA-2445 | wzm (O8) | AB010150 | 79.41 | TCACACCCATTGTTTATGTACTGAATTCATTACCTGC |
| OA-2446 | wzt (O8) | AB010150 | 79.73 | GCCGATAAACAAAATCAGTCCATTAAACAAGTTGAGCATA |
| OA-2801 | wzy (O11) | Laboratory stock | 80.91 | CGTTCAAGGTGGCAATTATATATTTCCATTGGTCACACTG |
| OA-2695 | wzy (O11) | Laboratory stock | 79.51 | CAGATGGAGTGTTTATGTATGTTCATTTATGCTAGGGGTA |
| OA-2700 | wzx (O15) | AY647261 | 76.6 | GAGTCATTGGTGTATCGAATTTTGGTGATCTGAGTTTTTC |
| OA-2701 | wzx (O15) | AY647261 | 81.53 | GCAATAAGTCAGGGTGCCAATTACCTACTGCCATTATTAA |
| OA-2452 | wzy (O25) | Laboratory stock | 79.73 | ATCCAGAACTTAACGATGTTAGTAGGCATTGTGATTAGTG |
| OA-2802 | wzx (O25) | Laboratory stock | 80.99 | AAATTAAGCCATGCAAGTAGTTTTACAGCGTCATATGCAG |
| OA-2453 | wzx (O27) | Laboratory stock | 79.55 | TCCTGTGCTATTTATGGGTTAGTTCTGATCAATTAACCTT |
| OA-2454 | wzy (O27) | Laboratory stock | 82.2 | TTGCTCTGTTCATAAAAGGCATTAGCACTTATTATGTCGT |
| OA-2697 | wzx (O78) | Laboratory stock | 79.23 | TCTTTTATCACATTGATTGGTGTTTGTTTTCTCTACCCAA |
| OA-2698 | wzy (O78) | Laboratory stock | 79.03 | TTATGAAAGGCTAACTGTTTACTTCGAATTTTCTCATGCT |
| OA-2666 | wzy (O85) | Laboratory stock | 80.13 | TTTCAGTACGTTAACTTTTGGTTGAGTGATGAACAACGTA |
| OA-2667 | wzy (O85) | Laboratory stock | 79.54 | AGTATTAACTCGTTTAGAAACCTTACAAGCTGGGAATGAT |
| OA-2806 | wzx (O114) | AY573377 | 80.82 | TCATAGGAAAGGATTAGAACATTGCTACAAGTGGTGGATT |
| OA-2809 | wzy (O114) | AY573377 | 79.78 | GGATGGAATGTTAATGGGTTATTTATTTCAGAAGCATGGG |
| OA-2670 | wzy (O115) | Laboratory stock | 79.02 | CAGTTTAGATGTTGTCCGATGGATTAATATAACGCTGTTT |
| OA-2671 | wzy (O115) | Laboratory stock | 80.79 | AGGCAGAAGGATGTTTGCTGTTATTTAATTGTATGCATGT |
| OA-2674 | wzy (O126) | Laboratory stock | 79.22 | ACGTAGTATTCTAATAATCGTGCTAACAATATGTGCGCTA |
| OA-2675 | wzy (O126) | Laboratory stock | 79.59 | TGGCATCTAAAATTATAAGTTCGTTAGGATTAGTGGCGAT |
| OA-2703 | wzx (O128) | AY217096 | 79.21 | GCCCATTGCATTCCTAAATTTGAAATGATTAATGCTATCC |
| OA-2460 | wzy (O128) | AY217096 | 82.02 | GCTAGGTATTTAGCAAATTCAACAGATTTGGCTGACTTTG |
| OA-2707 | wzx (O139) | DQ109552 | 79.35 | GGATTTCAGGGCCAATATTTTATGAGTTTTGTAGCCTTAT |
| OA-2708 | wzy (O139) | DQ109552 | 81.03 | ATGGAACCGTATGTACAATACTTTATAATCATGGGCCTGG |
| OA-2464 | wzy (O148) | DQ167407 | 79.01 | GCAATATTTGATACGTTAAGGGTTTATCTTTTCTCGGGAT |
| OA-2811 | wzy (O148) | DQ167407 | 80.22 | CAATGAGCAATATTTCGTACCATTAAGTGCAACAACCTTG |
| OA-2676 | wzx (O149) | DQ868764 | 79.96 | TATGGTATGCAATTACTGATTCATTAAGATTTGGAGGCGT |
| OA-2677 | wzx (O149) | DQ868764 | 79.89 | CGGTGCAAAGTTAATTCCGCTAACGATAATATGTTGTTTT |
| OA-2466 | wzy (O159) | EU294176 | 79.13 | GTTATAATGACAGTAGATTCAATCCTTTTCTTGGGTTGCA |
| OA-2812 | wzx (O159) | EU294176 | 80.08 | GCACTGAGCTATTTGGGTGTTAATTATTATGGTGTATGGA |
| OA-2680 | wzx (O166) | Laboratory stock | 79.39 | GCATGATGGTTTATTTTAGAGTGGATCGGTATTTTGTTGA |
| OA-2816 | wzy (O166) | Laboratory stock | 79.64 | TAGGAACAATAGTTTCGTTTCGAGATATAAGCGTTGATCG |
| OA-2684 | wzx (O167) | EU296408 | 79.35 | GCCATATATACTTCTGCAAATAAAATATTACAGGCGGCTC |
| OA-2685 | wzx (O167) | EU296408 | 79.2 | ACCGCAGTTGTTAATATCAGTAGTGGCAGTATATATCATT |
| OA-2689 | wzx (O173) | Laboratory stock | 79.31 | AGCTGTACTAATGTCATCATACACGATGGTAATTGGTATT |
| OA-2691 | wzy (O173) | Laboratory stock | 79.58 | TCTTAGAAAAGTTAGAGTTCCACCTCTTTTAGCATTGTGT |
| OA-2467 | rfpB (S. dysenteriae type 1) | S73325 | 79.16 | TGGGAGAAGAAAGTTATAAGTTAGCAAGAGAAAGATTCGA |
| OA-2468 | rfpB (S. dysenteriae type 1) | S73325 | 79.86 | AATTTATTATACTTGGCGCTATAGATAAGGAAAACCCCGG |
| OA-2469 | elt | S60731 | 79.8 | ACACATTAAGAATCACATATCTGACCGAGACCAAAATTGAb |
| OA-2470 | elt | S60731 | 80.16 | GCAAAAGAGAAATGGTTATCATTACATTTAAGAGCGGCGb |
| OA-2471 | estAp | M25607 | 79.74 | TTATCTTTCCCCTCTTTTAGTCAGTCAACTGAATCACTTGb |
| OA-2817 | estAp | M25607 | 76.7 | CGTTTAACTAATCTCAAATATCCGTGAAACAACATGACGG |
| OA-2473 | estAh | AY342059 | 76.5 | GTAGCAATTACTGCTGTGAATTGTGTTGTAATCCTGCTTG |
| OA-2819 | estAh | AY342059 | 78.5 | TTTCACCTTTCGCTCAGGATGCTAAACCAGTAGAGTCTTC |
| OA-2475 | estB | AY028790 | 79.11 | TTCTATTGCTACAAATGCCTATGCATCTACACAATCAAAb |
| OA-2476 | estB | AY028790 | 78.1 | AGGTTTTTTAGGGGTTAGAGATGGTACTGCTGGAGCATGb |
| OA-1993 | 16S rRNA | X80725 | 71.9 | TTGTACACACCGCCCGTCACACCAT |
| wl_4006 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT |
Tm was predicted using OligoArray software, version 2.0, except for probes OA-2700, OA-2817, OA-2473, OA-2819, and OA-1993, for which Tm was predicted using Primer Premier, version 5.0.
Probe used in our previous study (12).
DNA microarray preparation.
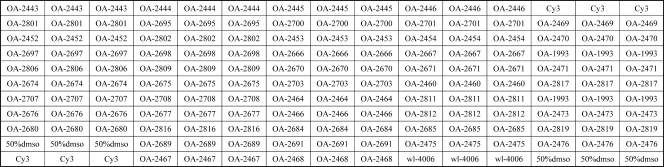
The probes were dissolved in 1× spotting buffer (3 M betaine, 3× SSC [0.45 M NaCl plus 0.045 M sodium citrate]) to a final concentration of 1 μg/μl and were printed on aldehyde group-modified glass slides (CapitalBio Corporation, Beijing, China) using the SpotArray 72 system (Perkin-Elmer Corporation, Waltham, MA). Each probe was spotted in triplicate. The printed slides were dried for 24 h at room temperature and were then cross-linked using a UV cross-linker (UVP Corporation, Upland, CA). The microarray slides were prehybridized at 45°C for 1 h in 100 ml of prehybridization buffer (containing 25 ml of 20× SSC, 10% sodium dodecyl sulfate [SDS], and 10 mg/ml bovine serum albumin [BSA]), washed twice in 0.1× SSC for 5 min each time, washed in Milli-Q water for 30 s, dried, and stored at room temperature in the dark. A schematic diagram of the probe positions on the microarray is shown in Fig. 1.
FIG. 1.
Schematic diagram of the microarray, showing the positions of immobilized probes spotted within a single well. Cy3 is a fluorescent dye. dmso, dimethyl sulfoxide. The sequences of the probes immobilized at each location are shown in Table 2.
Hybridization procedure.
A 40-μl aliquot of labeled PCR product was incubated at 65°C until dry; then it was resuspended in 20 μl of hybridization buffer (30% formamide, 5× SSC, 0.1% SDS, 0.001% salmon sperm DNA). After denaturation at 95°C for 5 min, 20 μl of the labeled target DNA was hybridized with the probes at 45°C for 16 h. After hybridization, the slide was washed once with solution A (2× SSC, 0.1% SDS) for 5 min, twice with solution B (0.1× SSC, 0.1% SDS) for 5 min each time, once with solution C (0.1× SSC) for 4 min, and finally once with solution D (0.01× SSC) for 15 s. The slide was then dried under a gentle air stream before it was scanned. For each DNA sample, at least three independent hybridization reactions were carried out to verify the reproducibility of the microarray method.
Data acquisition and analysis.
The hybridized microarray was scanned with a laser at 532 nm using the GenePix personal 4100A microarray scanner (Axon Instruments, Union City, CA), and the signals were calculated using GenePix Pro software, version 6.0. The data were analyzed, and the results were reported using the Bactarray Analyzer software (version 1.0) developed in-house.
Nucleotide sequence accession numbers.
The DNA sequences of the E. coli O25 and O27 O-antigen gene clusters have been deposited in the GenBank database under accession numbers GU014554 and GU014555, respectively.
RESULTS
O-antigen gene clusters within the E. coli serogroups of interest.
The O-antigen gene cluster sequences of E. coli serogroups O6, O8, O15, O114, O115, O126, O128, O139, O148, O149, O159, O167, and O173 were retrieved from the GenBank database (Table 2). The O-antigen gene cluster sequences of E. coli serogroups O11, O78, O85, and O166 were determined previously in our lab (unpublished data), and the sequences of E. coli serogroups O25 and O27 were obtained in this study. DNA sequencing between the galF and gnd genes was carried out for strains belonging to these two serogroups, and 17,566 bp (14 open reading frames [ORFs]) and 9,510 bp (7 ORFs) of sequence were obtained, respectively. The functions of each ORF in these O-antigen gene clusters were predicted on the basis of homology by searching the available databases.
Specific genes used for the detection and serotyping of ETEC.
Four genes that have been reported as virulence genes and used for the identification of ETEC, elt, estAp, estAh, and estB, were used as ETEC-specific genes in this study. The wzx and wzy genes have been reported as O-serogroup-specific genes in many studies and were therefore considered specific to each of the 19 serogroups targeted: O6, O8, O11, O15, O25, O27, O78, O85, O114, O115, O126, O128, O139, O148, O149, O159, O166, O167, and O173 (Table 2). The rfpB gene was used to differentiate E. coli serogroup O148 from S. dysenteriae type 1, because the wzx and wzy genes share almost 99% identity in these two serogroups (10).
Random PCR amplification method.
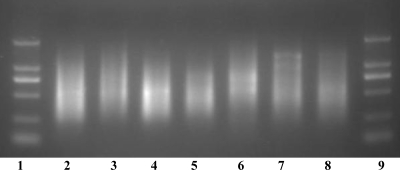
To amplify one or two O-serogroup-specific genes and four ETEC-specific virulence genes simultaneously in the ETEC strains belonging to the 19 targeted serogroups, a random PCR amplification method was used. A partially degenerate primer (9 bp at the 3′ end) was used in the first step of the procedure to randomly amplify fragments of DNA covering the entire genome, and a “tag” (17 bp at the 5′ end) was added to the amplified DNA at the same time. In the second step, a primer (with the same sequence as the tag) was used to further amplify the DNA synthesized in the first step, resulting in an exponential increase in the number of DNA molecules covering the whole genome. The amplification resulted in a DNA smear of fragments ranging from 250 to 2,000 bp (Fig. 2). The amplified DNA was then labeled with Cy3 in the third step to be used for hybridization with the probes printed on the microarrays.
FIG. 2.
Agarose gel electrophoresis of the random PCR products amplified from E. coli strains and S. dysenteriae type 1. Lanes 1 and 9, DNA markers of 100 bp, 250 bp, 500 bp, 750 bp, 1 kb, and 2 kb; lane 2, E. coli O6 (G1062); lane 3, E. coli O8 (G1602); lane 4, E. coli O11 (G1650); lane 5, E. coli O15 (G1201); lane 6, E. coli O114 (G1088); lane 7, E. coli O139 (G1208); lane 8, S. dysenteriae type 1 (G1018).
Specificity of the DNA microarray.
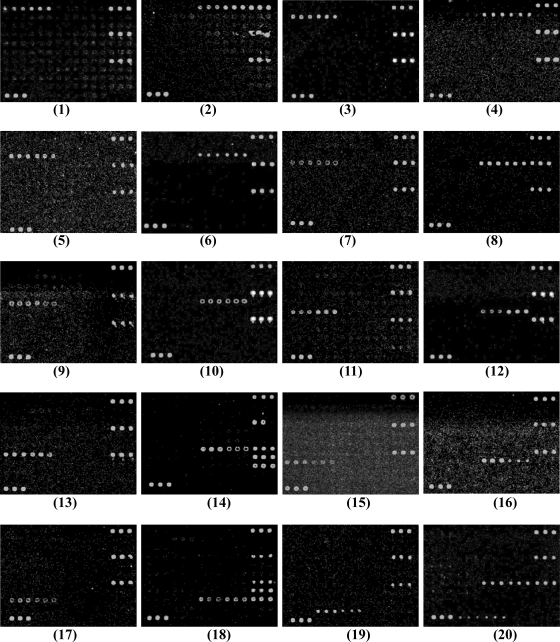
The DNA microarray was tested using 28 reference strains belonging to the 19 targeted E. coli O serogroups, 150 reference strains belonging to other E. coli O serogroups, and 23 reference strains of other, closely related species, including 13 Shigella strains and 10 Salmonella strains (Table 1). Through hybridization reactions with multiple strains from different sources representing each of the targeted serogroups, other serogroups, and other species, 51 specific probes were selected for inclusion in the microarray. These included 40 probes for O-serogroup-specific genes, 8 probes for virulence genes, 1 probe as a positive control, 1 probe as a negative control, and 1 probe as a positional reference and printing control (Table 2). All of the strains belonging to the 19 targeted serogroups or carrying virulence genes consistently hybridized to their corresponding probes with 100% specificity, indicating that the probes are effective at detecting their corresponding targeted serogroups and virulence genes. The hybridization results are shown in Fig. 3. For S. dysenteriae type 1, probes for the O148 wzy gene, the rfpB gene, the positive control, and the printing control hybridized to the microarray. For strains belonging to nontargeted serogroups of E. coli, and for Shigella and Salmonella strains, only the positive control and the printing control hybridized. A few of the E. coli strains belonging to nontargeted serogroups hybridized with probes to the elt gene, and none of the serogroup-specific probes hybridized (data not shown).
FIG. 3.
Microarray differentiation of reference strains belonging to different E. coli O serogroups and S. dysenteriae type 1. Panels: 1, E. coli O6 (G1062); 2, E. coli O8 (G1602); 3, E. coli O11 (G1657); 4, E. coli O15 (G1201); 5, E. coli O25 (G1111); 6, E. coli O27 (G1286); 7, E. coli O78 (G1235); 8, E. coli O85 (G1160); 9, E. coli O114 (G1088); 10, E. coli O115 (G1695); 11, E. coli O126 (G1679); 12, E. coli O128 (G1095); 13, E. coli O139 (G1658); 14, E. coli O148 (G1127); 15, E. coli O149 (G1061); 16, E. coli O159 (G1108); 17, E. coli O166 (G1216); 18, E. coli O167 (G1185); 19, E. coli O173 (G1093); 20, S. dysenteriae type 1 (G1018).
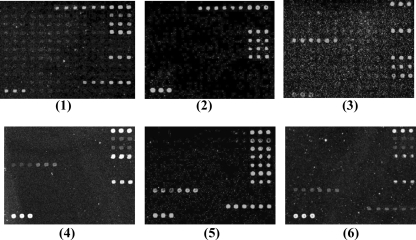
Double-blind test to verify the microarray.
A double-blind test was performed in order to verify the stability and specificity of the microarray. The test was carried out with 22 clinical isolates (Table 1) that had been characterized for O serotypes with specific antisera and for virulence genes by conventional PCR techniques at the Federal Institute for Risk Assessment (BfR) in Berlin, Germany. The hybridization patterns for representative clinical isolates are shown in Fig. 4. The detection results obtained with the microarray were consistent with the results obtained by conventional methods, indicating that the microarray assay is specific and reliable.
FIG. 4.
Microarray differentiation of some clinical isolates of E. coli. Panels: 1, E. coli O8 (G2493); 2, E. coli O8 (G2533); 3, E. coli O114 (G1336); 4, E. coli O114 (G1362); 5, E. coli O149 (G2525); 6, E. coli O149 (G2524).
Detection sensitivity of the microarray.
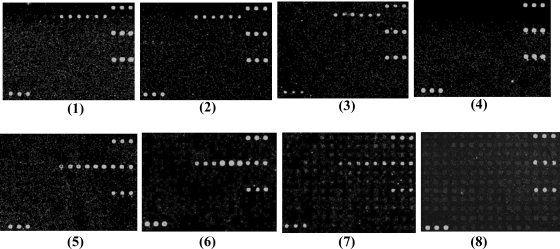
Serial dilutions of genomic DNAs (500, 100, 50, and 10 ng) of E. coli serogroup O15 strain G1383 and serogroup O85 strain G1160 were used as probes to test the sensitivity of the microarray. Strong hybridization signals were observed at DNA levels of 50 ng or higher (Fig. 5). We selected 50 ng of DNA as the most appropriate probe concentration for microarray detection. E. coli serogroup O15 strain G1383 was also serially diluted from 101 to 108 CFU/ml. Positive hybridization signals were obtained at 108 CFU/ml. By using 50 ng of DNA or 108 CFU/ml, all of the reference strains and clinical strains belonging to the 19 targeted serogroups could be detected correctly (data not shown).
FIG. 5.
Sensitivity of the microarray for the detection of genomic DNA from E. coli O15 strain G1383 at 500 ng (panel 1), 100 ng (panel 2), 50 ng (panel 3), and 10 ng (panel 4) and from E. coli O85 strain G1160 at 500 ng (panel 5), 100 ng (panel 6), 50 ng (panel 7), and 10 ng (panel 8).
DISCUSSION
Molecular methods for the detection of ETEC have been developed; however, these methods are based on PCR amplification of enterotoxin genes, so subsequent serotyping methods are required in order to fully characterize strains. Current serotyping methods for ETEC involve antiserum agglutination tests. Separate detection and serotyping methods increase the time and work required for pathogen identification. To our knowledge, this study is the first to develop a molecular assay for the parallel analysis of enterotoxin and serotyping genes in ETEC.
Information on the O serogroup of strains is a good indicator of strain variability and has been widely accepted as an epidemiological marker for the pathogenicity of E. coli strains. For example, serogroups O6, O8, O78, and O128 account for about half of the 988 ETEC isolates from 18 different countries (31). O serogroup differences among E. coli strains are almost entirely due to genetic variations in their O-antigen gene clusters, which include three groups of genes: nucleotide sugar synthesis genes, glycosyltransferase genes, and O-antigen-processing genes, including wzx and wzy. The wzx and wzy genes are usually specific to individual serogroups. PCR assays targeting these genes for the detection of pathogenic E. coli strains belonging to serogroups O157, O111, O123, and O86 have been reported (1, 9, 27, 28). In order to obtain all the wzx and wzy genes for ETEC serotyping, the O-antigen gene clusters of E. coli serogroups O25 and O27 were sequenced (the sequences of the O-antigen gene clusters of other ETEC serogroups were available through previous studies or from unpublished data generated by our lab).
DNA microarray technology is a relatively new methodology with many potential applications, one of which is the rapid and sensitive detection of bacterial pathogens. In comparison with traditional and PCR-based methods, microarrays offer the potential for high-throughput, specific, sensitive data collection, and microarray analysis has been successfully applied to the molecular typing of pathogenic microorganisms such as Streptococcus pneumoniae (29), group B streptococci (30), and Shigella (16). Two different amplification strategies, multiplex PCR and random amplification, have been reportedly used in these molecular typing studies to prepare DNA for hybridization to DNA microarrays. Multiplex PCR has been the standard method most commonly employed. However, multiplex PCR has several disadvantages vis-à-vis random amplification. First, the number of multiplex PCR primer pairs is limited because of cross-reactivity and primer-primer interactions. On the other hand, random amplification provides a highly comparable analysis but is not limited with regard to the number of genes that can be targeted; it is limited only with regard to the throughput of probes in the microarray. Also, random amplification does not require optimization of the primers themselves or of the quantity of primers used in the PCR, reducing the complexity and cost of amplification. Second, multiplex PCR can cause large amplification skews, which increase the risk of false-negative results, especially when the concentration of the template DNA is low. Random PCR, in contrast, is a relatively unbiased method and provides a more uniform genetic locus representation. Thus, random PCR is more effective than multiplex PCR at amplifying many gene locations. Third, to expand the spectrum of pathogens that can be detected by a multiplex PCR assay, extensive changes and reoptimization of the whole amplification procedure are likely required, whereas with random PCR, it is easy to add probes to the microarray in order to expand the detection spectrum without changing the steps prior to hybridization. In summary, compared with multiplex PCR, random PCR allows for amplification with minimal bias, providing an effective alternative for detecting pathogens using DNA microarrays. However, despite its advantages, random amplification is less sensitive than amplification via multiplex PCR. Still, the lower sensitivity of this method (50 ng of genomic DNA or 108 CFU/ml of organisms in pure culture) was not a problem in this case, because the strains used could easily be cultured to the required concentration.
The efficient detection of pathogenic microorganisms is crucial for the prevention and effective treatment of disease and, in some cases, for the safety of the wider community. The development of efficient and accurate detection methods is therefore of the utmost importance. The DNA microarray developed in this study has been shown to provide high-throughput, specific, and reliable detection and serotyping of ETEC. This approach has promising applications in clinical diagnosis and epidemiological surveillance. The strategy of using random PCR makes it easy to expand the detection range of the microarray by including more pathogens and/or serogroups; this will be the focus of future studies.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (30900255, 30788001, and 30870070), the National 863 Program (2007AA02Z106 and 2007AA021303), the National 973 Program (2009CB522603), and the National Key Programs for Infectious Diseases of China (2008ZX10004-002, 2008ZX10004-009, 2009ZX10004-108, 2008ZX10003, and 2008ZX10001-004).
Footnotes
Published ahead of print on 29 March 2010.
REFERENCES
- 1.Beutin, L., Q. Wang, D. Naumann, W. Han, G. Krause, L. Leomil, L. Wang, and L. Feng. 2007. Relationship between O-antigen subtypes, bacterial surface structures and O-antigen gene clusters in Escherichia coli O123 strains carrying genes for Shiga toxins and intimin. J. Med. Microbiol. 56:177-184. [DOI] [PubMed] [Google Scholar]
- 2.Black, R. E. 1993. Epidemiology of diarrhoeal disease: implications for control by vaccines. Vaccine 11:100-106. [DOI] [PubMed] [Google Scholar]
- 3.Black, R. E. 1990. Epidemiology of travelers' diarrhea and relative importance of various pathogens. Rev. Infect. Dis. 12(Suppl. 1):S73-S79. [DOI] [PubMed] [Google Scholar]
- 4.Blanco, M., N. L. Padola, A. Kruger, M. E. Sanz, J. E. Blanco, E. A. Gonzalez, G. Dahbi, A. Mora, M. I. Bernardez, A. I. Etcheverria, G. H. Arroyo, P. M. Lucchesi, A. E. Parma, and J. Blanco. 2004. Virulence genes and intimin types of Shiga-toxin-producing Escherichia coli isolated from cattle and beef products in Argentina. Int. Microbiol. 7:269-276. [PubMed] [Google Scholar]
- 5.Coimbra, R. S., F. Grimont, P. Lenormand, P. Burguiere, L. Beutin, and P. A. Grimont. 2000. Identification of Escherichia coli O-serogroups by restriction of the amplified O-antigen gene cluster (rfb-RFLP). Res. Microbiol. 151:639-654. [DOI] [PubMed] [Google Scholar]
- 6.Dallas, W. S. 1990. The heat-stable toxin I gene from Escherichia coli 18D. J. Bacteriol. 172:5490-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eom, H. S., B. H. Hwang, D. H. Kim, I. B. Lee, Y. H. Kim, and H. J. Cha. 2007. Multiple detection of food-borne pathogenic bacteria using a novel 16S rDNA-based oligonucleotide signature chip. Biosens. Bioelectron. 22:845-853. [DOI] [PubMed] [Google Scholar]
- 8.Ericsson, C. D. 2003. Travellers' diarrhoea. Int. J. Antimicrob. Agents 21:116-124. [DOI] [PubMed] [Google Scholar]
- 9.Feng, L., W. Han, Q. Wang, D. A. Bastin, and L. Wang. 2005. Characterization of Escherichia coli O86 O-antigen gene cluster and identification of O86-specific genes. Vet. Microbiol. 106:241-248. [DOI] [PubMed] [Google Scholar]
- 10.Feng, L., A. V. Perepelov, G. Zhao, S. D. Shevelev, Q. Wang, S. N. Senchenkova, A. S. Shashkov, Y. Geng, P. R. Reeves, Y. A. Knirel, and L. Wang. 2007. Structural and genetic evidence that the Escherichia coli O148 O antigen is the precursor of the Shigella dysenteriae type 1 O antigen and identification of a glucosyltransferase gene. Microbiology 153:139-147. [DOI] [PubMed] [Google Scholar]
- 11.Gorbach, S. L., B. H. Kean, D. G. Evans, D. J. Evans, Jr., and D. Bessudo. 1975. Travelers' diarrhea and toxigenic Escherichia coli. N. Engl. J. Med. 292:933-936. [DOI] [PubMed] [Google Scholar]
- 12.Han, W., B. Liu, B. Cao, L. Beutin, U. Kruger, H. Liu, Y. Li, Y. Liu, L. Feng, and L. Wang. 2007. DNA microarray-based identification of serogroups and virulence gene patterns of Escherichia coli isolates associated with porcine postweaning diarrhea and edema disease. Appl. Environ. Microbiol. 73:4082-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huilan, S., L. G. Zhen, M. M. Mathan, M. M. Mathew, J. Olarte, R. Espejo, U. Khin Maung, M. A. Ghafoor, M. A. Khan, Z. Sami, et al. 1991. Etiology of acute diarrhoea among children in developing countries: a multicentre study in five countries. Bull. World Health Organ. 69:549-555. [PMC free article] [PubMed] [Google Scholar]
- 14.Kosek, M., C. Bern, and R. L. Guerrant. 2003. The global burden of diarrhoeal disease, as estimated from studies published between 1992 and 2000. Bull. World Health Organ. 81:197-204. [PMC free article] [PubMed] [Google Scholar]
- 15.Lang, A. L., Y. L. Tsai, C. L. Mayer, K. C. Patton, and C. J. Palmer. 1994. Multiplex PCR for detection of the heat-labile toxin gene and Shiga-like toxin I and II genes in Escherichia coli isolated from natural waters. Appl. Environ. Microbiol. 60:3145-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y., D. Liu, B. Cao, W. Han, Y. Liu, F. Liu, X. Guo, D. A. Bastin, L. Feng, and L. Wang. 2006. Development of a serotype-specific DNA microarray for identification of some Shigella and pathogenic Escherichia coli strains. J. Clin. Microbiol. 44:4376-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maynard, C., F. Berthiaume, K. Lemarchand, J. Harel, P. Payment, P. Bayardelle, L. Masson, and R. Brousseau. 2005. Waterborne pathogen detection by use of oligonucleotide-based microarrays. Appl. Environ. Microbiol. 71:8548-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peñaranda, M. E., D. G. Evans, B. E. Murray, and D. J. Evans, Jr. 1983. ST:LT:CFA/II plasmids in enterotoxigenic Escherichia coli belonging to serogroups O6, O8, O80, O85, and O139. J. Bacteriol. 154:980-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeves, P. R., and L. Wang. 2002. Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264:109-135. [PubMed] [Google Scholar]
- 21.Sergeev, N., M. Distler, S. Courtney, S. F. Al-Khaldi, D. Volokhov, V. Chizhikov, and A. Rasooly. 2004. Multipathogen oligonucleotide microarray for environmental and biodefense applications. Biosens. Bioelectron. 20:684-698. [DOI] [PubMed] [Google Scholar]
- 22.Shivaswamy, S., and V. R. Iyer. 2007. Genome-wide analysis of chromatin status using tiling microarrays. Methods 41:304-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spicer, E. K., and J. A. Noble. 1982. Escherichia coli heat-labile enterotoxin. Nucleotide sequence of the A subunit gene. J. Biol. Chem. 257:5716-5721. [PubMed] [Google Scholar]
- 24.Stacy-Phipps, S., J. J. Mecca, and J. B. Weiss. 1995. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J. Clin. Microbiol. 33:1054-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenutz, R., A. Weintraub, and G. Widmalm. 2006. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev. 30:382-403. [DOI] [PubMed] [Google Scholar]
- 26.Urban, R. G., E. M. Pipper, L. A. Dreyfus, and S. C. Whipp. 1990. High-level production of Escherichia coli STb heat-stable enterotoxin and quantification by a direct enzyme-linked immunosorbent assay. J. Clin. Microbiol. 28:2383-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, L., H. Curd, W. Qu, and P. R. Reeves. 1998. Sequencing of Escherichia coli O111 O antigen gene cluster and identification of O111 specific genes. J. Clin. Microbiol. 36:3182-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, L., and P. R. Reeves. 1998. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect. Immun. 66:3545-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, Q., M. Wang, F. Kong, G. L. Gilbert, B. Cao, L. Wang, and L. Feng. 2007. Development of a DNA microarray to identify the Streptococcus pneumoniae serotypes contained in the 23-valent pneumococcal polysaccharide vaccine and closely related serotypes. J. Microbiol. Methods 68:128-136. [DOI] [PubMed] [Google Scholar]
- 30.Wen, L., Q. Wang, Y. Li, F. Kong, G. L. Gilbert, B. Cao, L. Wang, and L. Feng. 2006. Use of a serotype-specific DNA microarray for identification of group B Streptococcus (Streptococcus agalactiae). J. Clin. Microbiol. 44:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolf, M. K. 1997. Occurrence, distribution, and association of O and H serogroups, colonization factor antigens, and toxins of enterotoxigenic Escherichia coli. Clin. Microbiol. Rev. 10:569-584. [DOI] [PMC free article] [PubMed] [Google Scholar]