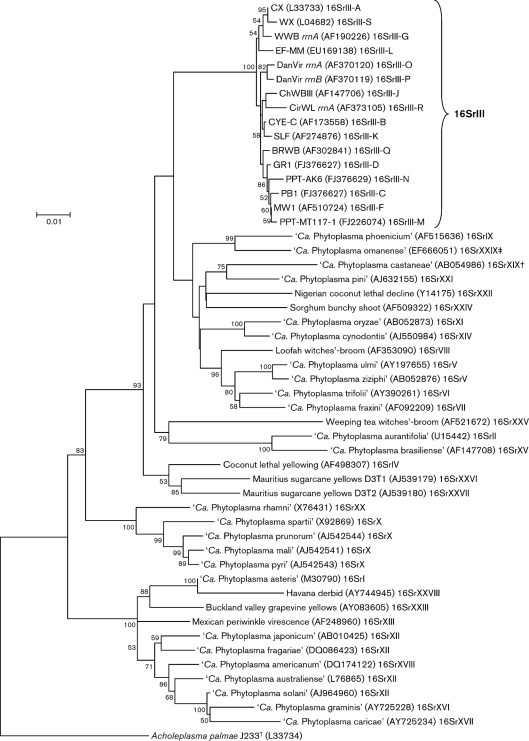
Fig. 4.
Phylogenetic tree inferred from analysis of 16S rRNA gene sequences. Minimum-evolution analysis was conducted using the close neighbour interchange (CNI) algorithm implemented in mega4 (Tamura et al., 2007). The initial tree for the CNI search was obtained by the neighbour-joining method. The reliability of the analysis was subjected to a bootstrap test with 1000 replicates. The taxa used in the phylogenetic tree reconstruction included reference strains of each phytoplasma 16Sr group, reference strains of subgroups belonging to the peach X-disease phytoplasma group and reference strains of each ‘Ca. Phytoplasma’ species (‘Ca. Phytoplasma allocasuarinae’ and ‘Ca. Phytoplasma lycopersici’ were not included because the available 16S rRNA gene sequences did not encompass the entire F2nR2 region. Reference strains of subgroups 16SrIII-E, 16SrIII-H and 16SrIII-I were not included because complete sequence information is not available). The sequence of Acholeplasma palmae J233T served as an outgroup during phylogenetic tree reconstruction. Bar, 0.01 nucleotide substitutions per site. †In the report by Jung et al. (2002), ‘Ca. Phytoplasma castaneae’ was assigned to group VI according to DNA sequence similarity, rather than results from RFLP analysis. In accordance with the more widely accepted RFLP-based classification system, this phytoplasma was reassigned to group 16SrXIX by Wei et al. (2007). ‡The original reference (Al-Saady et al., 2008) reported ‘Ca. Phytoplasma omanense’ as the reference member of a novel group designated group 16SrXIX. However, the group number 16SrXIX had been published previously (Wei et al., 2007) to accommodate a different phytoplasma, ‘Ca. P. castaneae’. Therefore, we assign ‘Ca. P. omanense’ to a new group, 16SrXXIX, subgroup 16SrXXIX-A.