Abstract
Over the past decade, the flavohaemoglobin Hmp has emerged as the most significant nitric oxide (NO)-detoxifying protein in many diverse micro-organisms, particularly pathogenic bacteria. Its expression in enterobacteria is dramatically increased on exposure to NO and other agents of nitrosative stress as a result of transcriptional regulation of hmp gene expression, mediated by (at least) four regulators. One such regulator, NsrR, has recently been shown to be responsible for repression of hmp transcription in the absence of NO in Escherichia coli and Salmonella, but the roles of other members of this regulon in Salmonella, particularly in surviving nitrosative stresses in vitro and in vivo, have not been elucidated. This paper demonstrates that an nsrR mutant of Salmonella enterica Serovar Typhimurium expresses high levels of Hmp both aerobically and anaerobically, exceeding those that can be elicited in vitro by supplementing media with S-nitrosoglutathione (GSNO). Elevated transcription of ytfE, ygbA, hcp and hcp is also observed, but no evidence was obtained for tehAB upregulation. The hyper-resistance to GSNO of an nsrR mutant is attributable solely to Hmp, since an nsrR hmp double mutant has a wild-type phenotype. However, overexpression of NsrR-regulated genes other than hmp confers some resistance of respiratory oxygen consumption to NO. The ability to enhance, by mutating NsrR, Hmp levels without recourse to exposure to nitrosative stress was used to test the hypothesis that control of Hmp levels is required to avoid oxidative stress, Hmp being a potent generator of superoxide. Within IFN-γ-stimulated J774.2 macrophages, in which high levels of nitrite accumulated (indicative of NO production) an hmp mutant was severely compromised in survival. Surprisingly, under these conditions, an nsrR mutant (as well as an nsrR hmp double mutant) was also disadvantaged relative to the wild-type bacteria, attributable to the combined oxidative effect of the macrophage oxidative burst and Hmp-generated superoxide. This explanation is supported by the sensitivity in vitro of an nsrR mutant to superoxide and peroxide. Fur has recently been confirmed as a weak repressor of hmp transcription, and a fur mutant was also compromised for survival within macrophages even in the absence of elevated NO levels in non-stimulated macrophages. The results indicate the critical role of Hmp in protection of Salmonella from nitrosative stress within and outside macrophages, but also the key role of transcriptional regulation in tuning Hmp levels to prevent exacerbation of the oxidative stress encountered in macrophages.
INTRODUCTION
The ability of Salmonella enterica serovar Typhimurium (S. typhimurium) to survive and proliferate within innate immune cells such as macrophages is central to its capacity to cause disease. The S. typhimurium genome encodes many mechanisms that allow resistance to the stressful environment encountered within the macrophage. Such stresses include the production by NAD(P)H oxidase (Phox) of superoxide anion (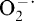 ) and other reactive oxygen species (ROS), and by inducible nitric oxide (NO) synthase (iNOS) of NO and other reactive nitrogen species (RNS) (Vazquez-Torres & Fang, 2001a; Fang, 2004).
) and other reactive oxygen species (ROS), and by inducible nitric oxide (NO) synthase (iNOS) of NO and other reactive nitrogen species (RNS) (Vazquez-Torres & Fang, 2001a; Fang, 2004).
The production of ROS, particularly 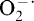 , in the univalent reduction of O2 by Phox is often referred to as the oxidative burst, and is thought to be activated around 1 h after infection (Tsolis et al., 1995). On activation of the phagocyte by uptake of Salmonella, the membrane and cytosolic components assemble in the cytoplasmic membrane in a process involving phosphorylation and cytoskeletal elements (Vazquez-Torres & Fang, 2001a; Forman & Torres, 2002). The
, in the univalent reduction of O2 by Phox is often referred to as the oxidative burst, and is thought to be activated around 1 h after infection (Tsolis et al., 1995). On activation of the phagocyte by uptake of Salmonella, the membrane and cytosolic components assemble in the cytoplasmic membrane in a process involving phosphorylation and cytoskeletal elements (Vazquez-Torres & Fang, 2001a; Forman & Torres, 2002). The 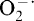 produced damages iron–sulfur [Fe–S] clusters and other targets (Imlay, 2002), whilst the hydrogen peroxide (H2O2) arising from
produced damages iron–sulfur [Fe–S] clusters and other targets (Imlay, 2002), whilst the hydrogen peroxide (H2O2) arising from 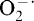 dismutation oxidizes protein thiols and [Fe–S] clusters and can create carbonyl and methionine sulfoxide adducts in proteins, thiols and membranes (Imlay, 2002). Furthermore, Fe(II) (formed, for example, by the reduction of Fe(III) by
dismutation oxidizes protein thiols and [Fe–S] clusters and can create carbonyl and methionine sulfoxide adducts in proteins, thiols and membranes (Imlay, 2002). Furthermore, Fe(II) (formed, for example, by the reduction of Fe(III) by 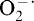 anion) reacts with H2O2 to generate the hydroxyl radical, which is a sufficiently powerful oxidant to react with virtually all organic molecules, including nucleic acids. To protect against oxidative stress, S. typhimurium possesses several antioxidant mechanisms (Farr & Kogoma, 1991; Vazquez-Torres & Fang, 2001b) that include not only superoxide dismutases (SODs) and hydroperoxidases, but also a type III secretory system that interferes with the trafficking of vesicles containing Phox to the phagosome (Fang & Vazquez-Torres, 2002).
anion) reacts with H2O2 to generate the hydroxyl radical, which is a sufficiently powerful oxidant to react with virtually all organic molecules, including nucleic acids. To protect against oxidative stress, S. typhimurium possesses several antioxidant mechanisms (Farr & Kogoma, 1991; Vazquez-Torres & Fang, 2001b) that include not only superoxide dismutases (SODs) and hydroperoxidases, but also a type III secretory system that interferes with the trafficking of vesicles containing Phox to the phagosome (Fang & Vazquez-Torres, 2002).
The production of RNS by macrophages is mediated by iNOS (Vazquez-Torres et al., 2000). Nitric oxide synthases (NOS) produce NO by the oxidation of a nitrogen atom of l-arginine to NO, using O2 and reducing equivalents provided by NAD(P)H as substrates. iNOS, the form of NOS most associated with antimicrobial activity, is primarily regulated at the transcriptional level and can be stimulated by the presence of microbial products and cytokines such as TNFα, IL-1 and IFN-γ (Vazquez-Torres et al., 2000; Kalupahana et al., 2005). Work by Chakravortty et al. (2002) demonstrated that the Salmonella pathogenicity island 2 (SPI-2) secretion system is able to interfere with the localization of iNOS and therefore aid avoidance of RNS. The extensive increase in production of RNS by macrophages following infection, often called the nitrosative burst, is thought to begin at around 8 h after infection in mouse macrophages (Eriksson et al., 2003), but may begin much earlier in human macrophages (Stevanin et al., 2002).
The protein considered most important in the detoxification of NO by S. typhimurium in aerobic conditions is the flavohaemoglobin Hmp. Flavohaemoglobins are the best-characterized class of microbial globin. They comprise two domains, a globin domain with a non-covalently bound haem B and a flavin domain with recognizable binding sites for FAD and NAD(P)H (Wu et al., 2003). Hmp was first identified in Escherichia coli (Vasudevan et al., 1991) and now has a clearly defined role in NO biology in that organism: its synthesis is markedly upregulated by NO (Poole et al., 1996), and hmp knockout mutants of E. coli and S. typhimurium are severely compromised for survival in the presence of NO in vitro (Crawford & Goldberg, 1998b; Membrillo-Hernandez et al., 1999). Salmonella Hmp has also been implicated in response to NO in human macrophages (Stevanin et al., 2002). In the presence of molecular O2, Hmp catalyses an oxygenase (Gardner et al., 1998) or denitrosylase (Hausladen et al., 1998) reaction in which NO is stoichiometrically converted to 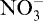 (Gardner et al., 1998), which is relatively innocuous. Extensive studies of the purified protein (e.g. Mills et al., 2001) have revealed some details of the reaction mechanism.
(Gardner et al., 1998), which is relatively innocuous. Extensive studies of the purified protein (e.g. Mills et al., 2001) have revealed some details of the reaction mechanism.
Regulation of Hmp levels in response to NO and related species in E. coli is complex. Control occurs predominantly at the transcriptional level and was shown in earlier studies to involve Fnr (Poole et al., 1996; Cruz-Ramos et al., 2002) and MetR (Membrillo-Hernandez et al., 1998). Recent computational and experimental studies have also implicated NsrR [product of the yjeB (nsrR) gene] in hmp regulation (Rodionov et al., 2005; Bodenmiller & Spiro, 2006). NsrR is an NO-sensitive transcriptional regulator of hmp and other genes known to be involved in nitrosative stress tolerance. It is a member of the Rrf2 family of transcriptional regulators, which also includes the IscR regulator that is involved in regulation of genes involved in [Fe–S] cluster biogenesis (Schwartz et al., 2001). Based on the similarity of NsrR to IscR, which contains an [Fe–S] cluster (Schwartz et al., 2001), and other members of the Rrf2 family (discussed in Rodionov et al., 2005), it has been suggested (Bodenmiller & Spiro, 2006) that NsrR contains an NO-sensitive [Fe–S] cluster. Very recently, a similar conclusion has been reached for NsrR in Bacillus subtilis (Nakano et al., 2006) and a role for NsrR in S. typhimurium hmp regulation has been proposed (Bang et al., 2006). In Salmonella, the response of hmp transcription to 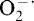 (generated by addition to cells of paraquat) is mediated by a further regulator, RamA (Hernandez-Urzua et al., 2007). There has been some confusion over the possible role of the ferric uptake regulation (Fur) protein in hmp regulation. Crawford & Goldberg (1998a) proposed that the iron-responsive regulator, Fur, represses hmp transcription and that this repression is lifted by NO on inactivation of Fur. Although these results have been retracted (Crawford & Goldberg, 2006), and others have suggested that Fur is not involved in Salmonella hmp expression (Bang et al., 2006), other promoters including hmp are controlled by nitrosylation of the Fur iron (D'Autreaux et al., 2002). Furthermore, we have recently published evidence based on newly constructed hmp–lacZ fusions and immunoblotting that Fur is a repressor of hmp transcription in both E. coli and Salmonella, albeit a weak one (Hernandez-Urzua et al., 2007). Given the global importance of Fur in intracellular iron management, a fur mutant of S. typhimurium might be compromised in its ability to resist killing within macrophages; conversely the modest upregulation of Hmp in the absence of Fur might confer a selective advantage.
(generated by addition to cells of paraquat) is mediated by a further regulator, RamA (Hernandez-Urzua et al., 2007). There has been some confusion over the possible role of the ferric uptake regulation (Fur) protein in hmp regulation. Crawford & Goldberg (1998a) proposed that the iron-responsive regulator, Fur, represses hmp transcription and that this repression is lifted by NO on inactivation of Fur. Although these results have been retracted (Crawford & Goldberg, 2006), and others have suggested that Fur is not involved in Salmonella hmp expression (Bang et al., 2006), other promoters including hmp are controlled by nitrosylation of the Fur iron (D'Autreaux et al., 2002). Furthermore, we have recently published evidence based on newly constructed hmp–lacZ fusions and immunoblotting that Fur is a repressor of hmp transcription in both E. coli and Salmonella, albeit a weak one (Hernandez-Urzua et al., 2007). Given the global importance of Fur in intracellular iron management, a fur mutant of S. typhimurium might be compromised in its ability to resist killing within macrophages; conversely the modest upregulation of Hmp in the absence of Fur might confer a selective advantage.
The main aim of this study was to investigate the roles of members of the NsrR regulon, including Hmp, in surviving nitrosative stresses in vitro and in vivo. The ability to enhance, by mutating NsrR, Hmp intrabacterial levels without recourse to exposure to nitrosative stress also allowed us to test the hypothesis that NsrR plays a key role in tuning Hmp levels, since we have previously demonstrated that, in vitro, Hmp is a potent generator of the products of partial oxygen reduction (Membrillo-Hernandez et al., 1996; Wu et al., 2004). In this paper, we report roles in vitro and in vivo for components of the NsrR regulon in S. typhimurium determined by studying the phenotypes of nsrR and hmp mutants, and of an nsrR hmp double mutant in which all regulon components other than Hmp are upregulated. We also re-examine the role of Fur and demonstrate that a fur mutant is compromised for survival within macrophages even in the absence of elevated NO levels in non-stimulated macrophages.
METHODS
Bacterial strains, media and growth conditions.
All studies were performed using wild-type Salmonella enterica Serovar Typhimurium ATCC 14028s or SL1344 and their isogenic derivatives (Table 1). Bacteria were cultured at 37 °C, with cultures shaking at 220 r.p.m., in LB broth (Sambrook et al., 1989) containing kanamycin (final concentration, 50 μg ml−1) or chloramphenicol (Cm) (final concentration, 25 μg ml−1) where appropriate. Media were inoculated at 1 % (v/v) with an overnight culture unless stated otherwise. For assessing the sensitivity of strains to methyl viologen (paraquat), H2O2 (both from Sigma) and S-nitrosoglutathione (GSNO), antibiotics were omitted from media. For assessing viability, cultures were spotted on nutrient agar (Sigma) containing the appropriate antibiotics. GSNO was synthesized according to the method of Hart (1985) and kindly donated by M. N. Hughes, University College London, UK. NO was synthesized according to the method in Poole et al. (1996).
Table 1.
Bacterial strains, phage and plasmids used in this study
| Strain, phage or plasmid | Genotype | Source/reference |
|---|---|---|
| S. typhimurium | ||
| ATCC 14028s | Wild-type | Crawford & Goldberg (1998b) |
| ATCC 14028s hmp | hmp : : kanr | Crawford & Goldberg (1998b) |
| ATCC 14028s fur | fur : : kanr | Kehres et al. (2002) |
| ATCC 14028s nsrR | nsrR : : cat | This study |
| ATCC 14028s nsrR hmp | nsrR : : cat, hmp : : kanr | This study |
| SL1344 | Xyl hisG rpsL | |
| SL1344 hcp hcr (nipAB) | SL1344 Δhcp hcr : : kanr | Kim et al. (2003) |
| SL1344 ytfE (nipC) | SL1344 ΔytfE : : kanr | Kim et al. (2003) |
| SL1344 nsrR hcp hcr | SL1344 nsrR : : cat, Δhcp hcr : : kanr | This study |
| SL1344 nsrR ytfE | SL1344 nsrR : : cat, ΔytfE : : kanr | This study |
| Bacteriophage | ||
| P22 | Lab stock | |
| Plasmids | ||
| pTP223 | λred, Tetr | J. Green, University of Sheffield |
| Poteete & Fenton (1984) | ||
| pBR322 | Apr Tetr | Lab stock |
| pBR322nsrR+ | Apr | This study |
To assess sensitivity to GSNO (3 mM) and paraquat (500 μM), growth curves were determined for cells grown in the absence or presence of the compounds in 10 ml LB in 250 ml flasks with side arms. Growth was measured by culture turbidity using a Klett–Summerson photoelectric colorimeter (Klett Manufacturing Co.) equipped with a no. 66 (red) filter. For H2O2 sensitivity assays, cultures were grown in 25 ml LB in 100 ml conical flasks for 3 h prior to addition of 5 mM H2O2. Sensitivity to H2O2 was determined by comparison of viable cell counts immediately prior to H2O2 addition with those at 10 and 25 min following addition. Viable counts were determined by conventional methods.
For Western blots, cells were grown as follows: anaerobic cultures were grown in six 8 ml glass tubes, filled to the brim with LB and sealed with Suba seals (Fisher); a glass ball in each tube facilitated resuspension of cells that had settled during static incubation. Aerobically grown cells were grown in six 10 ml LB batches in 250 ml flasks with side-arm for each strain. When the cultures reached mid-exponential phase (5 h anaerobic growth and 3 h aerobic growth, corresponding to Klett ∼40 and ∼80, respectively), 1 mM GSNO (final concentration) was added to three of the tubes/flasks for each strain. Anaerobic additions were made through the Suba seal using a Hamilton syringe. Cells were incubated for a further 2 h before being pooled and harvested by centrifugation at 5000 g for 10 min, 4 °C. Cells were stored as pellets at −20 °C.
Western blotting.
Pelleted cells were washed in 1 ml 50 mM Tris/HCl (pH 7.5), centrifuged at 13 800 g for 3 min and resuspended in 1 ml 50 mM Tris/HCl (pH 7.5). Over ice, each suspension was sonicated for 3×30 s with 1 min rests, at an amplitude of 10 μm. The disrupted cell suspension was centrifuged at 13 800 g and then the supernatant fractions were assayed for protein with the Bio-Rad protein assay kit and BSA as the standard. A sample (10 μg protein) of each sample was subjected to SDS-PAGE. Anti-Hmp antibody (Stevanin et al., 2000) was diluted 2000-fold for use with a 2000-fold dilution of peroxidase-conjugated monoclonal anti-rabbit immunoglobulin G (γ-chain specific, clone RG-96, A-1949; Sigma) as the secondary antibody. Western blots were carried out as described by Renart & Sandoval (1984); detection was done by using enhanced chemiluminescence (Amersham Biosciences).
Mutagenesis.
The λ Red system was used to promote replacement (first described in E. coli; Murphy, 1998) of a large portion of the nsrR gene with a Cm resistance (cat) gene. The cat gene from pACYC184 was PCR-amplified with primers having 40 bp of 5′ and 3′ flanking homology to the S. typhimurium nsrR gene. The linear DNA fragment was electroporated into wild-type S. typhimurium carrying pTP223 and transformants selected on nutrient agar containing Cm (final concentration 25 μg ml−1). Putative mutants were picked the following day and verified by PCR amplification of the nsrR region. The mutation was transduced into a clean wild-type background using P22 (Maloy et al., 1996), selecting for CmR.
Cloning.
The wild-type nsrR gene was amplified with primers engineered with terminal cut sites for EcoRI and BamHI restriction enzymes (at the 5′ and 3′ ends respectively). The PCR product was digested with the enzymes overnight at room temperature. pBR322 was simultaneously digested with the same enzymes at 4 °C. The restricted fragment and plasmid were then ligated with T4 DNA ligase (Promega), overnight at 4 °C. Ligation mixture (10 ng DNA) was used to transform competent DH5α cells (Invitrogen) and transformants were selected on nutrient agar containing 200 μg ampicillin ml−1. The recombinant plasmid was then reisolated using the Qiagen Qiaquick miniprep spin kit, and used to electroporate the mutant strain.
J774.2 macrophage culture and infection.
Macrophages for infection with Salmonella were cultured for 3 days in 24-well flat-bottom plates (2×105 cells per well) in Dulbecco’s Modified Eagle’s medium (DMEM) (D5796, Sigma) supplemented with 10 % fetal calf serum (FCS), at 37 °C in a humidified atmosphere containing 95 % air/5 % CO2. Where indicated, DMEM was supplemented with murine IFN-γ (RD Systems; 1000 U ml−1, final concentration) approximately 9 h prior to infection. For fluorescence microscopy, macrophages were cultured on sterile glass coverslips (BDH). Prior to infection, 12 ml LB was inoculated at 2 % with an overnight culture and cells incubated at 37 °C, shaking at 220 r.p.m., for ∼1.5 h or to an OD600 of ∼0.4, measured using a Jenway 6100 spectrophotometer, in cuvettes with a pathlength of 1 cm. Immediately prior to infection, bacteria were harvested by centrifugation for 3 min at 13 800 g and washed in phosphate-buffered saline, pH 7.4 (PBS). Bacteria were finally resuspended in DMEM+10 % FCS to give the appropriate c.f.u. ml−1, as determined by conventional methods.
Fluorescence microscopy.
Fluorescence microscopy was performed as described in Read et al. (1996). In brief, cells were fixed with 100 μl 2 % paraformaldehyde and stained using the nucleic acid stain 4,6-diamidino-2-phenylindole (DAPI), followed by a solution containing rabbit anti-Salmonella O antibody (Difco) diluted 1 : 50 in PBS. Cells were counterstained with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG antibody (Difco), diluted 1 : 20 in PBS. Coverslips were finally removed, dried, mounted in Vectashield (Molecular Probes), and viewed at a magnification of ×1000 in a DMRB 1000 fluorescence microscope (Leica). Internalization of bacteria was determined by subtracting the number of extracellular bacteria, identified by their co-localization with FITC-conjugated antibody, from the total number of bacteria exhibiting the DAPI stain.
Assay of intracellular Salmonella viability.
Macrophages were infected with Salmonella at an m.o.i. of 11.25 or 7.5, and incubated for 30 min at 37 °C in a humidified atmosphere (95 % air/5 % CO2) to allow internalization. Following incubation, wells were washed twice with PBS. At the time of sampling, J774.2 cells were lysed with 250 μl of 2 % sterile saponin for 20 min at 37 °C. Lysates were collected and wells washed with PBS (750 μl) to achieve maximum recovery of bacteria. The first samples were taken 30 min after infection, including three uninfected wells, after which the DMEM in the remaining wells was replaced with fresh DMEM containing gentamicin (100 μg ml−1) and incubated for a further 30 min to kill extracellular bacteria. The minimum bactericidal concentration for gentamicin of each strain was determined by conventional methods and was shown to be identical for all strains used. Subsequently, the media containing gentamicin at 100 μg ml−1 were replaced with media containing gentamicin at 25 μg ml−1 and incubated until lysis with saponin as described above. The cells were lysed at 30 min after initial infection, prior to gentamicin treatment, and at 4, 15 and 21 h after initial infection. All samples were taken in triplicate. Bacterial viability was determined using standard dilution techniques. Supernatants removed at 4, 15 and 21 h were stored at −21 °C for later nitrite and nitrate analyses.
Minimum bactericidal concentration test.
Cells were grown and washed exactly as they would be prior to macrophage infection, except that resuspension was in DMEM that contained various concentrations of gentamicin, up to 200 μg ml−1. Cells were incubated for 30 min before harvest, resuspended in 1 ml PBS and finally plated (10 μl spots) on nutrient agar and incubated overnight.
Assays of nitrite and nitrate accumulation in tissue culture supernatants.
Macrophage production of NO was measured by assaying 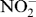 and
and 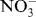 accumulation in culture supernatants as described in Stevanin et al. (2005) using a model 280i NO Analyser (Sievers).
accumulation in culture supernatants as described in Stevanin et al. (2005) using a model 280i NO Analyser (Sievers).
Quantitative real-time PCR (qRT-PCR).
RNA was extracted from 1 ml of a mid-exponential-phase aerobic culture of wild-type and nsrR strains using Qiagen RNAprotect and RNeasy kits as described by the manufacturer. Briefly, for each extraction, 1 ml culture was transferred into 2 ml RNAprotect reagent, followed by incubation at room temperature for 5 min. The samples were centrifuged at 6000 g for 10 min; the supernatant was discarded, and pellets frozen at −80 °C until use. The cell pellets were lysed after addition of 100 μl Tris-EDTA buffer containing lysozyme (Sigma) at a final concentration of 1 mg ml−1. RNA was purified as recommended by Qiagen and an on-column DNase digest was carried out using the RNase-free DNase set (Qiagen). RNA concentrations were determined spectrophotometrically using an Eppendorf Biophotometer. For cDNA synthesis, 4 μg RNA was added to 3 μl of a solution of random hexamer primers (Amersham Biosciences, 3 μg ml−1) and annealing was achieved by incubation at 65 °C for 10 min, 22 °C for 10 min and 2 min on ice. Reaction mixes (20 μl) containing 0.5 mM dATP, dTTP, dGTP and dCTP were incubated for 1 h at 42 °C with 200 units of Superscript II RNase-H Reverse Transcriptase (Invitrogen Superscript II kit). Following synthesis, cDNA was purified using a PCR purification kit (Qiagen) and eluted in 200 μl RNase-free water.
Gene-specific primers were designed to amplify 50- to 150- nucleotide fragments of target genes using PRIMER3 software (Rozen & Skaletsky, 2000). Each reaction was carried out in a total volume of 25 μl on a 96-well optical reaction plate (Applied Biosystems). Each well contained 0.5 μl 50× SYBR Green solution, 12.5 μl 2× Sensimix solution (Quantace), 3.25 pmol of each of the two primers and 5 μl cDNA sample. PCR amplification was carried out in an ABI 7700 thermocycler (PE Applied Biosystems) with the following thermal cycling conditions: 50 °C for 2 min; 95 °C for 10 min; 40 cycles of 95 °C for 15 s; 60 °C for 1 min. No-template reactions were included as negative controls. The data were analysed as described before (Flatley et al., 2005).
Reverse transcriptase PCR (RT-PCR).
cDNA was synthesized as described above; 1 μl of this was used as the template in a standard PCR reaction mixture using Accuzyme Polymerase (Bioline) to amplify.
Determination of inhibition of respiration by NO.
The method was adapted from Stevanin et al. (2000). Cultures (25 ml) were grown for 5.5 h. Cells were harvested by centrifugation and resuspended in 2 ml PBS. The buffer was saturated with air in a Clark-type polarographic oxygen electrode system (Rank Bros), comprising a water-jacketed (37 °C) Perspex chamber stirred magnetically; the membrane-covered electrode was situated at the bottom of the chamber below the stirrer. Cell suspension (300 μl) was added, a close-fitting lid applied to the chamber, and an ISO-NOP NO sensor (2 mm diameter) (WPI) was inserted through a custom-made capillary hole in the lid. Oxygen levels in the chamber were allowed to fall by 50–60 % through respiration before 100 μl of anoxic, NO-saturated solution was injected into the chamber using a Hamilton microsyringe. Oxygen consumption and NO levels were measured until the chamber contents became anaerobic.
Statistical analysis.
Parametric data were analysed for significance using the t-test and data plotted at means with error bars representing standard deviations (sd) or standard errors (sem) where stated. Non-parametric data were analysed for significance using the Wilcoxon signed rank test and data plotted as medians with error bars representing the 25th and 75th percentiles. Statistical significance was established at a P value of <0.05.
RESULTS
A S. typhimurium nsrR mutant constitutively synthesizes Hmp, aerobically and anaerobically
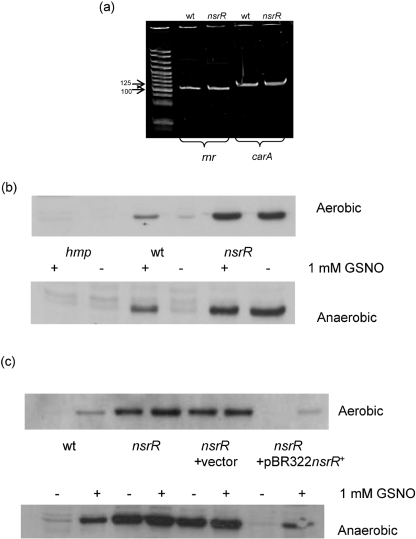
An nsrR mutant was created by the insertion of a Cm resistance cassette in S. typhimurium using the λ-red recombination system (Poteete & Fenton, 1984). The mutation was transduced into a wild-type background using phage P22 and confirmed by PCR amplification of the nsrR region. Immediately downstream of nsrR is a gene encoding riboendonuclease R, rnr, formerly vacB. In E. coli, nsrR and rnr are co-transcribed (Cairrao et al., 2003) and Rnr has been implicated in expression of virulence genes in Shigella and enteroinvasive E. coli (Tobe et al., 1992). Since there is a possibility that this is also the case in Salmonella, a complementation test was conducted as follows. pBR322 was engineered to carry the wild-type allele of nsrR (pBR322nsrR+) and the plasmid used to transform nsrR mutant by electroporation. To verify that the nsrR phenotype described is due to mutation of nsrR and not due to polar effects on the rnr gene, PCR was carried out on rnr cDNA from both wild-type and nsrR strains. The expression of rnr and of the gene encoding carbomoyl phosphatase, carA (as control), were indistinguishable in the wild-type strain and nsrR mutant (Fig. 1a).
Fig. 1.
rnr transcript and Hmp protein levels in Salmonella mutants. (a) rnr expression levels in wild-type and nsrR mutant strains. Cells were grown aerobically in LB and harvested during mid-exponential phase. PCR was performed on cDNA from these cells. Panel (a) is representative of an experiment carried out in duplicate. (b) Expression of Hmp in wild-type (wt), hmp and nsrR strains grown aerobically or anaerobically. (c) Expression of Hmp in wild-type (wt), nsrR, nsrR carrying pBR322 and nsrR carrying pBR322nsrR+ grown aerobically or anaerobically. Cells were exposed where indicated (+) to 1 mM GSNO in LB for 2 h during exponential phase, prior to harvesting. Cell-free extracts were resolved by SDS-PAGE and Western blots performed using antibody raised to E. coli purified Hmp. Similar protein loadings (10 μg) were used for each gel lane.
Wild-type, nsrR and hmp strains were grown to mid-exponential phase, both aerobically and anaerobically, in LB medium in the presence or absence of 1 mM GSNO. Western blots were performed using rabbit anti-E. coli-Hmp polyclonal antibodies as a probe (Fig. 1b, c) and clearly indicate the role of NsrR in the negative regulation of hmp in both aerobic and anaerobic conditions. An hmp mutant did not display a band corresponding to Hmp under any conditions. In the wild-type strain, a band corresponding to Hmp was detected only after cells were exposed to GSNO, whereas in the nsrR mutant strain a strong band corresponding to Hmp was observed in both the presence and absence of GSNO, illustrating loss of negative regulation in the mutant. Even in the presence of 1 mM GSNO, hmp was not maximally expressed in the wild-type strain, as demonstrated by comparison of the Hmp band intensity with that of the nsrR mutant. Our findings are in agreement with Rodionov et al. (2005), who used bioinformatics to predict that NsrR negatively regulates hmp and several other genes in a small regulon. Recently Bang et al. (2006) also identified NsrR as a regulator of Hmp in S. typhimurium using qRT-PCR. Western blots identical to those described above were also carried out on the nsrR strains carrying pBR322 and pBR322nsrR+ (Fig. 1c) and show that Hmp is expressed as in the nsrR mutant in nsrR carrying pBR322 and as in the wild-type in nsrR carrying pBR322nsrR+; this expression of nsrR+ in trans restores its function as a repressor of hmp under non-nitrosating conditions.
The S. typhimurium NsrR regulon contains at least four other genes
Rodionov et al. (2005) identified the following S. typhimurium genes as being potentially under the regulation of NsrR: the hcp-hcr (nipAB) operon, hmp, ytfE (nipC) and tehB. In addition they suggested that E. coli ygbA is also regulated by NsrR. We tested the influence of the nsrR mutation on the expression of these genes using qRT-PCR. In addition, we looked at the expression of tehA, which is located just upstream of tehB, and with which it may be co-transcribed. The upregulation of mRNA levels in the nsrR mutant strain in comparison to wild-type was as follows: ytfE, 544; hcp, 284; ygbA, 31.8; hcr, 4.1; tehA, 1.1; tehB, 1.1. Data presented here are representative of a single RT-PCR experiment using cDNA from wild-type and nsrR strains in technical triplicates. A biological repeat of this was carried out giving a similar trend in gene expression levels. carA was again used as the control gene.
The function of ytfE is unclear, although several global transcriptional analyses in E. coli have shown it to be highly induced under conditions of nitrosative stress (Mukhopadhyay et al., 2004; Justino et al., 2005; Pullan et al., 2007). The ygbA gene also has no known function. hcp and hcr are both upregulated in our nsrR mutant. In E. coli, they are considered to be co-transcribed (van den Berg et al., 2000), yet levels of hcr mRNA levels were only 4.1-fold greater in the mutant strain. This might be explained by differential stability of the mRNA transcript leading to faster degradation of the hcr portion of the transcript.
The hmp gene is the NsrR regulon member conferring GSNO resistance
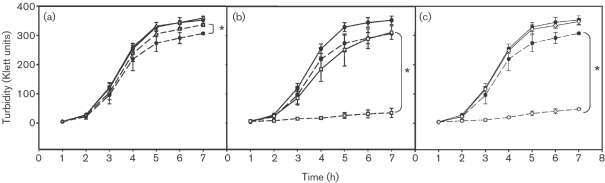
On finding that hmp is constitutively expressed in an nsrR mutant (Fig. 1b), we tested resistance of the mutant to nitrosative stress by growth in LB medium in the presence or absence of 3 mM GSNO. Fig. 2(a) shows that the aerobic growth characteristics of nsrR and wild-type strains are indistinguishable. In the presence of 3 mM GSNO, however, growth of the wild-type strain was more severely affected than that of the nsrR strain, presumably because enhanced expression of hmp in the nsrR strain affords more protection from GSNO. However, to establish if other members of the NsrR regulon protect against GSNO, an nsrR hmp double mutant was created by P22 transduction of the hmp mutation into the nsrR mutant, and the construct verified by Western blot analysis (not shown). Growth curves in the absence or presence of 3 mM GSNO (Fig. 2b) show that removal of Hmp from the nsrR mutant abrogates the enhanced GSNO resistance of the nsrR mutant. Fig. 2(c) shows that a single hmp mutant has the same growth profile as the nsrR hmp double mutant in the presence of 3 mM GSNO. Mutants in ytfE and hcp hcr were also tested for their sensitivity to GSNO and were found to display growth profiles similar to that of wild-type (data not shown). Thus, no other members of the NsrR regulon are directly involved in conferring the ability to grow in the presence of GSNO.
Fig. 2.
Hmp, but not other members of the NsrR regulon, confers growth tolerance to GSNO. Growth was in LB medium in the absence (solid lines) or presence (dashed lines) of 3 mM GSNO, measured turbidimetrically and shown as Klett units. (a) Comparison of wild-type (•) and nsrR (▵). (b) Comparison of wild-type (•) and nsrR hmp (□). (c) Comparison of wild-type (•) and hmp (◊). Data points are means±sd in three independent experiments. *, P=<0.05 at the 7 h data point using Student’s t-test.
Overexpression of genes other than hmp can protect S. typhimurium respiration from inhibition by NO
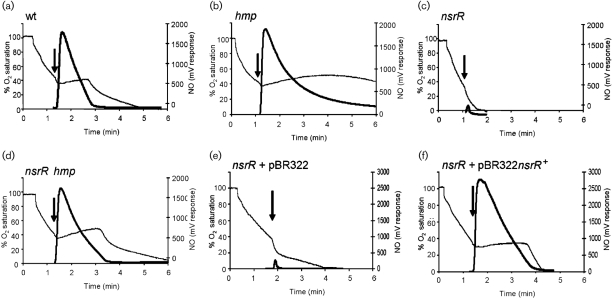
To test the effect of NO on respiration, mid-exponential-phase cells were allowed to respire aerobically in PBS until dissolved oxygen tension reached approximately 40 % of air saturation, at which point 100 μl NO-saturated solution was added to the cells. Fig. 3(a) shows that respiration of wild-type cells was totally but temporarily inhibited. NO addition was visualized by a rapid upward excursion of the NO electrode output while oxygen uptake was abruptly arrested for less than 2 min. After NO concentration fell, reaching negligible amounts approximately 1.5 min post-NO addition, oxygen uptake resumed at a rate similar to that before NO addition. The slight upwards deflection in the oxygen electrode traces seen after inhibition of respiration (and also in Fig. 3b and d) presumably reflects polarographic drift or the back-diffusion of oxygen into the chamber through the capillary used for NO addition. A trace similar to that of wild-type can be seen in Fig. 3(f), where the nsrR mutant is complemented by pBR322nsrR+. A slightly prolonged inhibition of oxygen uptake can be seen in this trace, presumably attributed to multiple copies of the nsrR gene exerting higher repression on hmp than the single copy of nsrR in the wild-type genome. In the hmp strain (Fig. 3b), although NO addition was evident by a similar spike to that seen in the wild-type experiment, the disappearance of NO was slower, and even after 6 min a significant amount of NO could be measured by the NO electrode. Oxygen uptake by the hmp mutant was severely affected by addition of NO and did not resume within 5 min of NO addition. Thus Hmp protects S. typhimurium from the effects of NO on aerobic respiration. Conversely, the nsrR mutant and the nsrR strain carrying pBR322, synthesizing high levels of Hmp, were unaffected by NO (Fig. 3c, e); indeed, NO was consumed so quickly in each case that added NO was virtually undetectable by the electrode. Furthermore, directly following NO addition, a transient increase in oxygen uptake occurred, attributable to the Hmp-catalysed oxygenase mechanism (Gardner et al., 1998). To assess the possible roles of other members of the NsrR regulon, the polarographic analyses were extended to the nsrR hmp mutant (Fig. 3d). In this strain, despite the lack of Hmp-catalysed NO removal, NO was consumed more rapidly than for the hmp mutant (Fig. 3b) and respiration resumed within 2 min of NO addition. We conclude that other gene(s) in the nsrR regulon, expressed to high levels in the nsrR hmp mutant, exert protective effects on respiration in the presence of NO. To try to establish which NsrR-regulated genes were responsible for protection in the nsrR hmp double mutants, mutants in ytfE and hcp hcr along with mutants in nsrR ytfE and nsrR hcp hcr were all tested for their ability to withstand NO inhibition of respiration. The ytfE and hcp hcr mutants showed profiles similar to that of wild-type (data not shown), and nsrR ytfE and nsrR hcp hcr mutants showed profiles similar to that of the nsrR mutant (data not shown). These results indicate that ytfE and hcp hcr do not have a role in protection of respiration from NO, when a functional hmp gene is still present. Several attempts were made throughout this study to construct an ygbA mutant to investigate its role in the above but were unsuccessful.
Fig. 3.
Hmp is the most significant member of the NsrR regulon conferring resistance of aerobic respiration to NO. Intact cells were allowed to respire in PBS until approximately 40 % of the dissolved oxygen remained, when 100 μl of an NO-saturated solution was injected. Fine line traces represent dissolved oxygen tension and thick lines represent NO. (a) Wild-type (wt); (b) hmp mutant; (c) nsrR mutant; (d) nsrR hmp mutant; (e) nsrR mutant carrying pBR322; (f) nsrR mutant carrying complementing plasmid. Data shown are representative of at least three independent experiments. Note that the recovery of respiration after NO inhibition in (a), (d) and (f) is spontaneous.
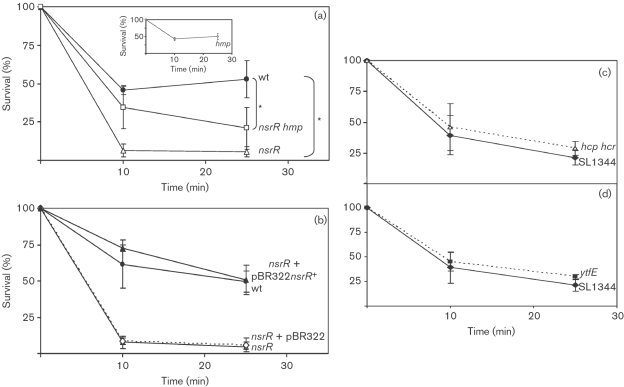
nsrR, hmp and fur mutations attenuate survival in J774.2 macrophages
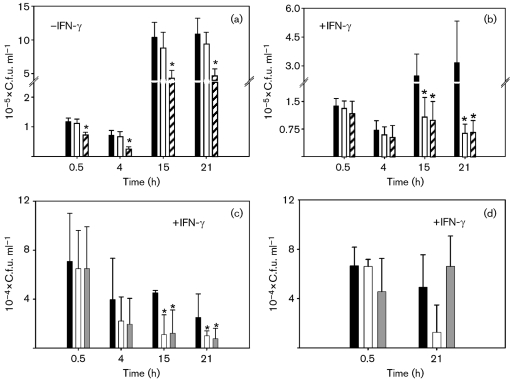
In this study we used the mouse macrophage cell line J774.2. Prior to investigation of intracellular survival, each Salmonella strain was examined for the extent of association (defined here as the total number of bacteria either bound or internalized per macrophage), binding and internalization with J774.2, via fluorescence microscopy. After 30 min incubation, all bacterial strains associated similarly with the macrophages (data not shown) so that subsequent viable counts reflect intracellular survival and not differences in internalization. We also carried out gentamicin sensitivity tests to demonstrate that all strains were affected equally by the antibiotic used to kill extracellular bacteria following 30 min infection: all strains were equally sensitive to gentamicin (data not shown). Furthermore, the effects of bacterial infection on survival of J774.2 cells over 21 h were also examined; a trypan blue assay revealed insignificant effects on cytotoxicity (data not shown). Following infection (mean m.o.i. of 11.25), macrophages were lysed after 0.5, 4, 15 and 21 h and viable counts were carried out on the lysates to enumerate surviving intracellular bacteria. Fig. 4(a) shows that, in the absence of prior IFN-γ stimulation, there was no significant difference in the intracellular survival of wild-type and hmp mutant strains. However, mutation in the global iron response regulator, Fur, significantly affected the intracellular survival and proliferation of bacteria; resulting in significantly lower bacterial numbers being recovered, even after the initial 0.5 h infection period, indicating that the fur strain is unable to survive even the initial exposure to the intracellular macrophage environment. After 4 h infection, the number of recovered wild-type cells was over threefold higher than fur cells, and after 15 and 21 h wild-type counts were over double that of fur mutant counts (Fig. 4a). These data suggest that the null fur mutant is severely attenuated in naïve macrophages, but that mutation in the NO-detoxifying globin gene, hmp, does not significantly affect the ability of S. typhimurium to survive under the same conditions. The data also suggest that fur is attenuated in these macrophages by an antimicrobial mechanism other than the nitrosative burst.
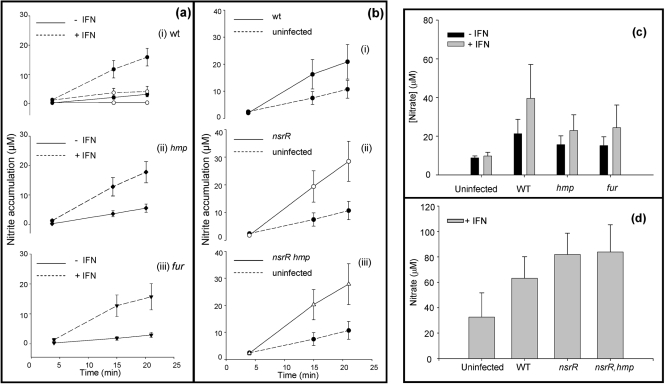
Fig. 4.
Intracellular survival of Salmonella is compromised in a fur mutant, but only in the presence of γ-IFN in an hmp mutant. Bacterial cells (∼2.5×106) suspended in 250 μl DMEM+10 % FCS were used to infect ∼2×105 J774.2 cells (a) and J774.2 cells stimulated with IFN-γ (b), giving an m.o.i. of ∼11. Bacterial cells (∼1.5×106) similarly suspended were used to infect ∼2×105 J774.2 cells (c), giving an m.o.i. of ∼7.5. The intracellular survival and proliferation was followed over time. At 0.5 h, cell counts include extracellular bacteria together with viable internalized bacteria. At subsequent time points, extracellular bacteria have been killed using gentamicin, leaving only viable intracellular bacteria. At each time point, counts for wild-type and for hmp and fur mutant strains are shown by the black, white and hatched bars, respectively in (a) and (b). In (c), at each time point, counts for wild-type and for nsrR and nsrR hmp mutant strains are shown by the black, white and shaded bars, respectively. In (d) counts for wild-type (wt), nsrR and nsrR carrying pBR322nsrR are shown by black, white and shaded bars, respectively. Results are medians±25th and 75th percentiles from between 6 and 12 separate experiments. *, P<0.05 using the Wilcoxon statistical analysis test.
In subsequent experiments, macrophages were incubated with IFN-γ (1000 U ml−1) for approximately 9 h prior to infection. IFN-γ enhances a nitrosative stress response in macrophages upon infection with S. typhimurium (Kalupahana et al., 2005). As expected (Liu et al., 1999), the numbers of bacteria recovered from activated J774.2 cells were lower than those recovered from non-stimulated J774.2 cells for all three strains (Fig. 4b), with wild-type numbers increasing only fivefold between 4 h and 21 h infection as compared to a 16-fold increase in non-activated cells (Fig. 4a). The enhanced nitrosative stress associated with activation severely impairs survival and proliferation of the hmp mutant. The greatest differences between wild-type and both hmp and fur cell counts were seen at 15 and 21 h after infection, with numbers of wild-type reaching approximately two- and fourfold that of the mutant strains, respectively. These results clearly indicate the requirement for Hmp and Fur in maximal survival and proliferation in activated macrophages.
Since the nsrR mutant is hyper-resistant to GSNO (Fig. 2a) and NO (Fig. 3c), we hypothesized that this mutant may display additional resistance to killing by macrophages. Kim et al. (2003) reported that, when hcp, hcr and ytfE (nipABC) Salmonella mutants were used to infect mice, the ability of animals to clear the infection with these strains was diminished at low doses. We compared the survival of both the nsrR and nsrR hmp mutants to wild-type bacteria in IFN-γ-stimulated macrophages (Fig. 4c) using a slightly lower m.o.i. of 7.5 to aid bacterial counting. Comparison of the wild-type strain in Fig. 4(b, c) indicates that a larger m.o.i. is required for proliferation of S. typhimurium in activated J774.2 cells over a 21 h period and that a decrease in m.o.i. (in this case of ∼33 %), does not correspond to an equal fall in c.f.u. ml−1 of wild-type Salmonella at 0.5 h (in this case it fell to ∼53 %), suggesting that J774.2 cells are more effective at limiting survival of Salmonella immediately after infection, and reducing subsequent proliferation, when the initial infection load is smaller. Our primary concern in this study was not to discern the correlation between infecting m.o.i. and survival of wild-type Salmonella but rather to assess whether there is a difference in the survival of wild-type and mutant strains of Salmonella. Surprisingly, we observed that both the nsrR and nsrR hmp mutants survived similarly intracellularly (Fig. 4c), with bacterial counts for both strains being twofold lower than wild-type at 15 and 21 h lysis time points. A possible explanation of these data is that Hmp at high levels exerts toxic effects by the production of 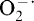 (Membrillo-Hernandez et al., 1996; Wu et al., 2004), so that the nsrR mutant, having elevated Hmp levels, is disadvantaged in macrophages due to exacerbated sensitivity to the oxidative response produced by the macrophages. To clarify that the nsrR mutation was not having polar effects on the downstream gene, rnr, which could have a role in virulence (Tobe et al., 1992), intracellular survival of the nsrR mutant carrying pBR322nsrR+ was assessed in activated macrophages. Fig. 4(d) shows that inclusion of wild-type nsrR on a plasmid caused the intracellular survival of the nsrR mutant to return to the same level as the wild-type strain at 21 h.
(Membrillo-Hernandez et al., 1996; Wu et al., 2004), so that the nsrR mutant, having elevated Hmp levels, is disadvantaged in macrophages due to exacerbated sensitivity to the oxidative response produced by the macrophages. To clarify that the nsrR mutation was not having polar effects on the downstream gene, rnr, which could have a role in virulence (Tobe et al., 1992), intracellular survival of the nsrR mutant carrying pBR322nsrR+ was assessed in activated macrophages. Fig. 4(d) shows that inclusion of wild-type nsrR on a plasmid caused the intracellular survival of the nsrR mutant to return to the same level as the wild-type strain at 21 h.
Nitrite and nitrate accumulation in tissue culture supernatants
We sought to verify the elevated production of NO in IFN-γ-stimulated J774.2 cells by assaying 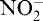 levels in tissue culture, as NO produced by iNOS is expected to be oxidized to
levels in tissue culture, as NO produced by iNOS is expected to be oxidized to 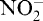 in the presence of oxygen (Ignarro et al., 1993; Wink et al., 1993; Kharitonov et al., 1994). In the presence of Hmp, NO is detoxified to
in the presence of oxygen (Ignarro et al., 1993; Wink et al., 1993; Kharitonov et al., 1994). In the presence of Hmp, NO is detoxified to 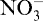 (Gardner et al., 1998; Hausladen et al., 1998). Supernatant fractions from non-stimulated macrophages accumulated very little
(Gardner et al., 1998; Hausladen et al., 1998). Supernatant fractions from non-stimulated macrophages accumulated very little 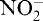 compared to their activated counterparts (Fig. 5a ii). In non-activated, uninfected cells,
compared to their activated counterparts (Fig. 5a ii). In non-activated, uninfected cells, 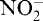 levels were ∼0.3 μM throughout the study, whereas in stimulated macrophages,
levels were ∼0.3 μM throughout the study, whereas in stimulated macrophages, 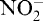 increased consistently throughout the experiment to ∼4 μM after 21 h, confirming activation of iNOS.
increased consistently throughout the experiment to ∼4 μM after 21 h, confirming activation of iNOS. 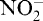 levels detected in non-activated J774.2 cells in the presence of wild-type (Fig. 5a i) and fur (Fig. 5a iii) strains were similar, with accumulations of around 3 μM after 21 h of infection whereas levels of
levels detected in non-activated J774.2 cells in the presence of wild-type (Fig. 5a i) and fur (Fig. 5a iii) strains were similar, with accumulations of around 3 μM after 21 h of infection whereas levels of 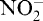 were slightly higher in hmp-infected J774.2 cells (5.5 μM) (Fig. 5a ii). This could reflect the accumulation of NO in hmp infected cells and its oxidation to
were slightly higher in hmp-infected J774.2 cells (5.5 μM) (Fig. 5a ii). This could reflect the accumulation of NO in hmp infected cells and its oxidation to 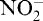 , whereas in wild-type and fur strains, Hmp converts NO to
, whereas in wild-type and fur strains, Hmp converts NO to 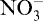 . We tested this hypothesis by measuring the accumulation of
. We tested this hypothesis by measuring the accumulation of 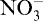 in tissue culture media over time. As with
in tissue culture media over time. As with 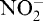 , steady increases in
, steady increases in 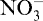 concentration were detected over the time-course of infection with all strains. Fig. 5(c) shows the total
concentration were detected over the time-course of infection with all strains. Fig. 5(c) shows the total 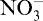 accumulated at 21 h.
accumulated at 21 h. 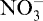 levels reached a mean of 8.7 μM in the supernatants of non-activated, uninfected cells after 21 h.
levels reached a mean of 8.7 μM in the supernatants of non-activated, uninfected cells after 21 h. 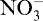 accumulation in wild-type-infected cells had a mean of 21.3 μM (Fig. 5c). Both hmp- and fur-infected J774.2 tissue culture supernatants showed a similar accumulation of
accumulation in wild-type-infected cells had a mean of 21.3 μM (Fig. 5c). Both hmp- and fur-infected J774.2 tissue culture supernatants showed a similar accumulation of 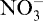 , which was significantly lower than that of wild-type bacteria at around 15 μM. However, IFN-γ-activated J774.2 cells infected with Salmonella produced large amounts of
, which was significantly lower than that of wild-type bacteria at around 15 μM. However, IFN-γ-activated J774.2 cells infected with Salmonella produced large amounts of 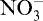 over 21 h (∼40 μM for the wild-type strain), compared to the non-stimulated, infected J774.2 cells. The
over 21 h (∼40 μM for the wild-type strain), compared to the non-stimulated, infected J774.2 cells. The 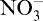 concentration in supernatants from stimulated macrophages was somewhat lower, at around 23 μM, for hmp- and fur-infected J774.2 cells (Fig. 5c). The lower level of
concentration in supernatants from stimulated macrophages was somewhat lower, at around 23 μM, for hmp- and fur-infected J774.2 cells (Fig. 5c). The lower level of 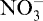 in the supernatant of hmp-infected cells could reflect an inability of the hmp mutant to detoxify NO to
in the supernatant of hmp-infected cells could reflect an inability of the hmp mutant to detoxify NO to 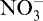 ; however, as the fur infected cells showed similar
; however, as the fur infected cells showed similar 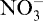 accumulation, it is probably more likely that differences in
accumulation, it is probably more likely that differences in 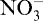 accumulation seen in both non-activated and activated tissue culture supernatants could be explained by the fact that iNOS activation may be infection-load dependent (Witthoft et al., 1998).
accumulation seen in both non-activated and activated tissue culture supernatants could be explained by the fact that iNOS activation may be infection-load dependent (Witthoft et al., 1998).
Fig. 5.
Nitrite and nitrate concentrations in supernatant fractions from intracellular killing assays. Tissue culture supernatants (DMEM+10 % FCS) were removed from macrophages immediately prior to lysis of macrophages. Panel (a) shows assay results for nitrite analysis in the absence and presence of IFN-γ for: (i) wild-type, (ii) hmp and (iii) fur bacteria. Uninfected J774.2 cells are shown by open circles in (i). Panel (b) shows results for nitrite analysis in the presence of IFN-γ for (i) wild-type, (ii) nsrR and (iii) nsrR hmp bacteria. Solid lines show the data for infected macrophages and dashed lines show the data for uninfected controls. Panel (c) shows nitrate analysis after 21 h for uninfected J774.2 cells and after infection with wild-type (wt), hmp and fur bacteria. Data are shown for the absence (black bars) and presence (grey bars) of IFN-γ. Panel (d) shows nitrate analysis after 21 h of uninfected J774.2 cells, wild-type (wt), nsrR and nsrR hmp bacteria. Concentrations of nitrite and nitrate represent the accumulation in fresh DMEM+10 % FCS media containing gentamicin (25 μg ml−1) that was applied to the macrophages immediately following 30 min incubation with DMEM+10 % FCS containing gentamicin (100 μg ml−1). Points and error bars represent means±sem of at least three experimental repeats.
Tissue culture supernatants were also collected from experiments where wild-type, nsrR and nsrR hmp strains were used to infect IFN-γ-stimulated macrophages and used to assay 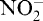 and
and 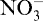 accumulations (Fig. 5b and d, respectively). Although nsrR mutant bacteria have elevated levels of Hmp relative to wild-type, no significant differences in
accumulations (Fig. 5b and d, respectively). Although nsrR mutant bacteria have elevated levels of Hmp relative to wild-type, no significant differences in 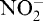 or
or 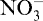 levels were detected, perhaps as a result of the loss of bacterial viability (see Fig. 4c) and therefore lower activity levels of iNOS (Witthoft et al., 1998). Macrophages infected with the nsrR hmp double mutant also showed no significant difference from the wild-type in
levels were detected, perhaps as a result of the loss of bacterial viability (see Fig. 4c) and therefore lower activity levels of iNOS (Witthoft et al., 1998). Macrophages infected with the nsrR hmp double mutant also showed no significant difference from the wild-type in 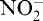 and
and 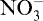 accumulation (Fig. 5b, d), perhaps due to the lack of significant NO-metabolizing activity by the strain and the poor viability of such bacteria (Fig. 4c).
accumulation (Fig. 5b, d), perhaps due to the lack of significant NO-metabolizing activity by the strain and the poor viability of such bacteria (Fig. 4c).
An nsrR mutation enhances sensitivity to oxidative stress, even in the absence of Hmp
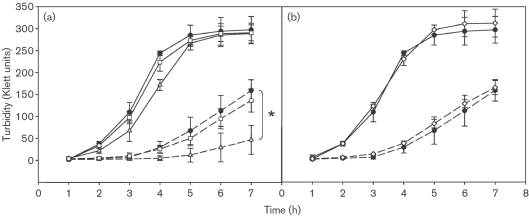
We hypothesized that the compromised survival of nsrR mutant bacteria in macrophages is due to intracellular oxidative stress resulting from excessive Hmp synthesis and its reduction of O2 to 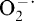 (Membrillo-Hernandez et al., 1996; Wu et al., 2004). This was tested in vitro using two different oxidative stress agents to study the response of wild-type, nsrR, hmp and nsrR hmp strains. Paraquat produces
(Membrillo-Hernandez et al., 1996; Wu et al., 2004). This was tested in vitro using two different oxidative stress agents to study the response of wild-type, nsrR, hmp and nsrR hmp strains. Paraquat produces 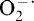 and is toxic to bacteria in vitro (Liochev & Fridovich, 1993; Halliwell & Gutteridge, 1999). In the absence of paraquat, all three strains grew to similar final densities although growth of the nsrR mutant, but not the double mutant, was significantly slower during most of the growth curve (Fig. 6a). This is consistent with an inhibitory effect of excess Hmp synthesis in the absence of NO. Growth of the nsrR mutant in the presence of 200 μM paraquat showed some deficiency when compared to wild-type (data not shown). However, in the presence of 500 μM paraquat, growth of the nsrR mutant was almost completely inhibited (Fig. 6a), whereas the nsrR hmp strain (Fig. 6a) had growth characteristics similar to the wild-type, as did the hmp mutant (Fig. 6b). These results demonstrate that Hmp, but not other products of the NsrR regulon, exacerbate paraquat sensitivity. The data also suggest that the poor survival of the nsrR hmp mutant in macrophages (Fig. 4c) is due not to oxidative stress, but to the inability of the mutant to deal with nitrosative stress, as in the hmp mutant.
and is toxic to bacteria in vitro (Liochev & Fridovich, 1993; Halliwell & Gutteridge, 1999). In the absence of paraquat, all three strains grew to similar final densities although growth of the nsrR mutant, but not the double mutant, was significantly slower during most of the growth curve (Fig. 6a). This is consistent with an inhibitory effect of excess Hmp synthesis in the absence of NO. Growth of the nsrR mutant in the presence of 200 μM paraquat showed some deficiency when compared to wild-type (data not shown). However, in the presence of 500 μM paraquat, growth of the nsrR mutant was almost completely inhibited (Fig. 6a), whereas the nsrR hmp strain (Fig. 6a) had growth characteristics similar to the wild-type, as did the hmp mutant (Fig. 6b). These results demonstrate that Hmp, but not other products of the NsrR regulon, exacerbate paraquat sensitivity. The data also suggest that the poor survival of the nsrR hmp mutant in macrophages (Fig. 4c) is due not to oxidative stress, but to the inability of the mutant to deal with nitrosative stress, as in the hmp mutant.
Fig. 6.
Sensitivity of an nsrR mutant to paraquat. Growth was measured in the absence (solid lines) or presence (dashed lines) of 500 μM paraquat. (a) Comparison of wild-type (•) with nsrR (▵) and nsrR hmp (□) mutants; (b) comparison of wild-type (•) with hmp mutant (◊). Data are represented as means±sd of at least three independent experiments. *, P<0.05 at the 7 h data point using Student’s t-test.
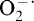 spontaneously dismutates in solution to give peroxide (Halliwell & Gutteridge, 1999; Imlay, 2002). We therefore also performed sensitivity tests to H2O2. Strains were grown for 3 h before addition of 5 mM H2O2. Culture samples were taken immediately prior to the addition and then at 10 and 25 min following the addition. Viable counts were compared to the counts immediately prior to the stress (Fig. 7). Approximately 50 % of wild-type cells survived following 10 and 25 min exposure to H2O2 (Fig. 7a). The hmp strain gave a profile almost identical to that of the wild-type (Fig. 7a inset). The most severely affected strain was the nsrR mutant, with less than 10 % of cells surviving at 10 and 25 min after H2O2 treatment (Fig. 7a). To confirm that the H2O2 sensitivity was a trait of the nsrR mutation and therefore overexpression of hmp, nsrR mutants carrying pBR322nsrR+ or pBR322 were also tested (Fig. 7b). The nsrR mutant carrying pBR322 gave a killing profile identical to the nsrR mutation whereas the nsrR mutant carrying pBR322nsrR+ behaved like the wild-type. However, the nsrR hmp mutant also exhibited exacerbated H2O2 sensitivity, compared to wild-type, particularly after 25 min (Fig. 7a). The reason for this result is unknown, although it seems possible that overexpression of a member of the NsrR regulon, other than Hmp, contributes to sensitivity to H2O2. Interestingly, recent studies in E. coli have shown that mutants in hcp are more sensitive to H2O2 (Almeida et al., 2006), implicating Hcp in the oxidative stress response; thus overexpression of hcp is unlikely to increase oxidative stress sensitivity of the nsrR hmp mutant. The Salmonella hcp hcr and ytfE mutants were not increased in their sensitivity to H2O2 in these experiments (Fig. 7c, d), indicating possible differences in the properties of Hcp in E. coli and Salmonella.
spontaneously dismutates in solution to give peroxide (Halliwell & Gutteridge, 1999; Imlay, 2002). We therefore also performed sensitivity tests to H2O2. Strains were grown for 3 h before addition of 5 mM H2O2. Culture samples were taken immediately prior to the addition and then at 10 and 25 min following the addition. Viable counts were compared to the counts immediately prior to the stress (Fig. 7). Approximately 50 % of wild-type cells survived following 10 and 25 min exposure to H2O2 (Fig. 7a). The hmp strain gave a profile almost identical to that of the wild-type (Fig. 7a inset). The most severely affected strain was the nsrR mutant, with less than 10 % of cells surviving at 10 and 25 min after H2O2 treatment (Fig. 7a). To confirm that the H2O2 sensitivity was a trait of the nsrR mutation and therefore overexpression of hmp, nsrR mutants carrying pBR322nsrR+ or pBR322 were also tested (Fig. 7b). The nsrR mutant carrying pBR322 gave a killing profile identical to the nsrR mutation whereas the nsrR mutant carrying pBR322nsrR+ behaved like the wild-type. However, the nsrR hmp mutant also exhibited exacerbated H2O2 sensitivity, compared to wild-type, particularly after 25 min (Fig. 7a). The reason for this result is unknown, although it seems possible that overexpression of a member of the NsrR regulon, other than Hmp, contributes to sensitivity to H2O2. Interestingly, recent studies in E. coli have shown that mutants in hcp are more sensitive to H2O2 (Almeida et al., 2006), implicating Hcp in the oxidative stress response; thus overexpression of hcp is unlikely to increase oxidative stress sensitivity of the nsrR hmp mutant. The Salmonella hcp hcr and ytfE mutants were not increased in their sensitivity to H2O2 in these experiments (Fig. 7c, d), indicating possible differences in the properties of Hcp in E. coli and Salmonella.
Fig. 7.
Percentage survival of cells after treatment with 5 mM H2O2. (a) Data are shown for the wild-type strain (•), the nsrR mutant (▵) and the nsrR hmp double mutant (□). (a) Inset, sensitivity of hmp mutant to peroxide stress. (b) Comparison of wild-type (•), nsrR mutant (▵), nsrR carrying pBR322 (dashed line) and nsrR carrying complementing plasmid (▴). (c, d) Sensitivity of hcp hcr (▵) and ytfE (▪) mutants, respectively, to H2O2 (dashed lines), and comparison with isogenic wild-type, SL1344 (⧫). Data are represented as means±sd of at least three independent experiments. *, P<0.05 at the 7 h data point using Student’s t-test.
DISCUSSION
This study has confirmed that NsrR is a major regulator of hmp in S. typhimurium both aerobically and anaerobically (Fig. 1b, c), and mutation in nsrR protects against the effects of GSNO in vitro. This protection is shown to be a direct consequence of overexpression of hmp, as removal of hmp in an nsrR mutant abrogates the extra protection and leads to a growth pattern, in the presence of GSNO, that is similar to that of a single hmp mutant (Fig. 2). The absence of NsrR, with consequent overexpression of Hmp, also protects respiration from inhibition by NO (Fig. 3). However, in contrast to the growth effects, we show that this is due, not only to overexpression of hmp, but also to the overexpression of other gene(s) since, in the nsrR hmp double mutant, respiration recovers in a manner similar to the wild-type strain. In an hmp single mutant, respiration does not recover over the time period of the experiment, presumably due to the failure of Hmp to detoxify NO. Thus, no other members of the NsrR regulon can protect growth from the adverse effects of nitrosative stress (GSNO) but one or more of the other members of the NsrR regulon (hcp, hcr, ytfE, ygbA or other unidentified gene) are involved in protection of the oxidative electron-transport chain from inhibition by NO, at least in the absence of functional hmp. tehA and tehB expression in the S. typhimurium nsrR mutant was not increased, whereas in E. coli tehA was shown to be weakly regulated by nsrR (Bodenmiller & Spiro, 2006). The tehA and tehB gene products are involved in resistance to tellurite and other toxic compounds (Turner et al., 1997) but roles for these genes in nitrosative stress resistance are unknown.
It has been suggested that ytfE may have a role in [Fe–S] cluster biogenesis in E. coli (Justino et al., 2006). In S. typhimurium, ytfE has been identified as having a promoter that is maximally induced by 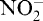 in a pH-dependent manner (Kim et al., 2003). Mutants in ytfE are less readily cleared by mice in low-dose infection, the mechanism of which remains unknown (Kim et al., 2003). Hcp contains two [Fe–S] clusters of unusual redox properties, and Hcr has sequence similarity to flavin-containing and [Fe–S]-containing class 1 NADH oxidoreductases and has been shown to reduce Hcp in the presence of NADH (van den Berg et al., 2000). Like ytfE, the hcp hcr genes have also been shown to be optimally expressed in 1 mM
in a pH-dependent manner (Kim et al., 2003). Mutants in ytfE are less readily cleared by mice in low-dose infection, the mechanism of which remains unknown (Kim et al., 2003). Hcp contains two [Fe–S] clusters of unusual redox properties, and Hcr has sequence similarity to flavin-containing and [Fe–S]-containing class 1 NADH oxidoreductases and has been shown to reduce Hcp in the presence of NADH (van den Berg et al., 2000). Like ytfE, the hcp hcr genes have also been shown to be optimally expressed in 1 mM 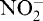 at pH 5, induced in NO-producing macrophages and less readily cleared by mice in low-dose infection (Kim et al., 2003). Recently, the E. coli Hcp has been implicated in the oxidative stress response (Almeida et al., 2006). The ygbA gene has no known function, although it has been shown to be regulated by nsrR in E. coli (Bodenmiller & Spiro, 2006).
at pH 5, induced in NO-producing macrophages and less readily cleared by mice in low-dose infection (Kim et al., 2003). Recently, the E. coli Hcp has been implicated in the oxidative stress response (Almeida et al., 2006). The ygbA gene has no known function, although it has been shown to be regulated by nsrR in E. coli (Bodenmiller & Spiro, 2006).
Despite an nsrR mutant being hyper-resistant to GSNO and NO in vitro (Figs 2 and 3), we show that tight regulation of hmp expression is of paramount importance for survival in murine macrophages (Fig. 4), where bacteria will experience a range of stresses. Preliminary experiments assessing the survival of hmp and fur mutants in naïve macrophages suggested that the nitrosative stress response in these macrophages is not strong enough to attenuate survival of the hmp mutant (Fig. 4a) but other stresses, particularly levels of radical and oxidizing species, have deleterious effects on the intracellular survival of a mutant in the global regulator, Fur (Fig. 4a). Previous work looking at the intracellular survival of a Salmonella SL1344 fur mutant in macrophages demonstrated that this strain was not severely attenuated in intracellular survival (Garcia-del Portillo et al., 1993). Later experiments by Wilmes-Riesenberg et al. (1996) also showed that the SL1344 fur mutant was not attenuated in its survival within J774.2 macrophages but that the degree to which a fur mutation affects virulence depends on the background strain of Salmonella, with the SL1344 fur mutant showing only a small increase in LD50 in mouse infection. These results could indicate why the 14028s fur strain used in this study shows an attenuation in J774.2 cells not seen before.
IFN-γ was used to enhance the antimicrobial nitrosative response. IFN-γ is the major macrophage-activating cytokine (Unanue, 1993). The resulting activation of iNOS is illustrated by the reduced survival of the hmp mutant (Fig. 4b), which was significantly lower than wild-type in activated cells. The fall in hmp bacterial counts demonstrates the impaired ability of this strain to withstand nitrosative stress due to lack of the NO-detoxifying globin. 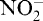 and
and 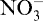 accumulation in tissue culture supernatants of infected cells confirms the enhanced activity of iNOS in the presence of IFN-γ (Fig. 5). Salmonella are known to interfere with the localization of iNOS (Chakravortty et al., 2002), which may be enough to defend against RNS damage in the non-stimulated J774.2 but, in activated cells, where greater amounts of RNS are produced, Hmp appears to be required. This is in agreement with Bang et al. (2006), who showed a role for Hmp in acute and chronic mouse infection models and also presented data showing that a hmp mutant is not attenuated in infection of mice fed with the iNOS inhibitor L-NIL. Generally, RNS production by macrophages is thought to cause bacterial cell death via (in)activation of enzymes, ion channels and transcription factors, and mutation of DNA by strand breakage (Szabo et al., 1996). However, a mechanism involving the IFN-γ-stimulated production of NO causing an inhibition of Salmonella SPI-2 effector expression has been reported, triggering the Salmonella-containing vacuole to interact more efficiently with compartments of the lysosomal/endosomal system, resulting in increased effectiveness of Salmonella killing by macrophages (McCollister et al., 2005).
accumulation in tissue culture supernatants of infected cells confirms the enhanced activity of iNOS in the presence of IFN-γ (Fig. 5). Salmonella are known to interfere with the localization of iNOS (Chakravortty et al., 2002), which may be enough to defend against RNS damage in the non-stimulated J774.2 but, in activated cells, where greater amounts of RNS are produced, Hmp appears to be required. This is in agreement with Bang et al. (2006), who showed a role for Hmp in acute and chronic mouse infection models and also presented data showing that a hmp mutant is not attenuated in infection of mice fed with the iNOS inhibitor L-NIL. Generally, RNS production by macrophages is thought to cause bacterial cell death via (in)activation of enzymes, ion channels and transcription factors, and mutation of DNA by strand breakage (Szabo et al., 1996). However, a mechanism involving the IFN-γ-stimulated production of NO causing an inhibition of Salmonella SPI-2 effector expression has been reported, triggering the Salmonella-containing vacuole to interact more efficiently with compartments of the lysosomal/endosomal system, resulting in increased effectiveness of Salmonella killing by macrophages (McCollister et al., 2005).
With enhanced expression of hmp and other genes in the NsrR regulon, it was predicted that the nsrR mutant might have an advantage over wild-type S. typhimurium in survival within IFN-γ-activated macrophages. However, our results suggest quite the opposite, both the nsrR and the nsrR hmp mutants being attenuated in IFN-γ-activated macrophages (Fig. 4c). Furthermore, in vitro work demonstrated that the nsrR mutant is hyper-sensitive to both paraquat (Fig. 6a) and H2O2 and that the nsrR hmp mutant is sensitive to H2O2 (Fig. 7). We have previously demonstrated that the presence of haem and FAD in Hmp not only provides a facile electron transfer from NAD(P)H to O2 and NO in the active site where 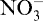 formation occurs, but also renders the protein susceptible to participation in additional redox chemistry. Specifically, reduction of oxygen to
formation occurs, but also renders the protein susceptible to participation in additional redox chemistry. Specifically, reduction of oxygen to 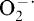 was first proposed by Orii et al. (1992), then demonstrated experimentally (Membrillo-Hernandez et al., 1996; Wu et al., 2004). Hmp acts as a reductase of broad specificity, reducing oxygen, cytochrome c and Fe(III) hydroxamate K, apparently without the involvement of the haem, since cytochrome c reduction can be demonstrated in the presence of CO (Poole et al., 1997). Recent work in S. typhimurium (Bang et al., 2006) has confirmed that the FAD-binding domain of Hmp mediates hyper-susceptibility to oxidative stress. Although these authors suggest specifically the reduction of Fe(III) to Fe(II) as demonstrated before (Poole et al., 1997), and subsequent Fenton reaction chemistry (Woodmansee & Imlay, 2003), the reductive abilities of Hmp might facilitate numerous other reactions. Fig. 8 illustrates the network of interactions centring on Hmp. A large corpus of biochemical and physiological data supports the concept that Hmp possesses critical O2-dependent NO-detoxifying activity yet also has the potential for generating ROS in the absence of NO. A multitude of layers of transcriptional regulation provide the fine-tuning of Hmp levels necessary. Furthermore, additional modes of protection from nitrosative stress are clearly indicated by the current results from the nsrR hmp construct. Further work is required to assess the role of other NsrR regulon members.
was first proposed by Orii et al. (1992), then demonstrated experimentally (Membrillo-Hernandez et al., 1996; Wu et al., 2004). Hmp acts as a reductase of broad specificity, reducing oxygen, cytochrome c and Fe(III) hydroxamate K, apparently without the involvement of the haem, since cytochrome c reduction can be demonstrated in the presence of CO (Poole et al., 1997). Recent work in S. typhimurium (Bang et al., 2006) has confirmed that the FAD-binding domain of Hmp mediates hyper-susceptibility to oxidative stress. Although these authors suggest specifically the reduction of Fe(III) to Fe(II) as demonstrated before (Poole et al., 1997), and subsequent Fenton reaction chemistry (Woodmansee & Imlay, 2003), the reductive abilities of Hmp might facilitate numerous other reactions. Fig. 8 illustrates the network of interactions centring on Hmp. A large corpus of biochemical and physiological data supports the concept that Hmp possesses critical O2-dependent NO-detoxifying activity yet also has the potential for generating ROS in the absence of NO. A multitude of layers of transcriptional regulation provide the fine-tuning of Hmp levels necessary. Furthermore, additional modes of protection from nitrosative stress are clearly indicated by the current results from the nsrR hmp construct. Further work is required to assess the role of other NsrR regulon members.
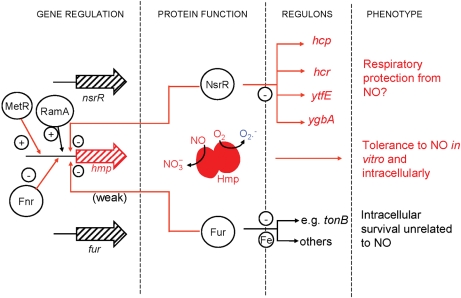
Fig. 8.
Contributions of NsrR, Hmp and Fur to NO tolerance and intracellular survival in Salmonella. Entities (genes, proteins, regulatory events, metabolic products) that increase on nitrosative stress are shown in red. Positive (+) and negative (−) gene regulation patterns refer to the effects of regulators in the absence of NO. Superoxide production from Hmp, which decreases with NO, is shown in blue. Transcription of hmp is positively regulated by MetR (in response to NO+-mediated homocysteine deprivation) and RamA (on exposure to superoxide), but negatively regulated by Fnr, Fur (weakly) and NsrR (strongly). Hmp catalyses the aerobic conversion of NO to nitrate, but (predominantly at the flavin domain) also reduces oxygen to superoxide in the absence of NO. In the presence of NO, repression by NsrR of hcp, hcr, ytfE and ygbA is lifted, and repression by Fur of at least 30 (Ratledge & Dover, 2000) genes is also lifted. At the right are shown the roles revealed in the present work of Hmp and the NsrR and Fur regulon components.
It is notable that, within the intracellular environment, the nsrR strain appears unable to withstand the oxidative stress emanating from both macrophage and Hmp, despite the possession by S. typhimurium of several antioxidant enzymes including four SODs, namely two periplasmic Cu, Zn-SODs (SodCI and SodCII) (Fang et al., 1999), a Mn-SOD (SodA) (Tsolis et al., 1995) and an Fe-SOD (SodB). SODs dismutate 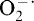 to H2O2, which can then be disproportionated into H2O and O2 by catalases. S. typhimurium also possesses two catalases, encoded by katG and katE. KatG and KatE are often referred to as HPI and HPII, respectively (for a review see Storz & Imlay, 1999). The results presented here suggest that these mechanisms of protection are overwhelmed by excess
to H2O2, which can then be disproportionated into H2O and O2 by catalases. S. typhimurium also possesses two catalases, encoded by katG and katE. KatG and KatE are often referred to as HPI and HPII, respectively (for a review see Storz & Imlay, 1999). The results presented here suggest that these mechanisms of protection are overwhelmed by excess 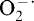 production in the nsrR mutant when Hmp is overexpressed. In vitro, the nsrR hmp double mutant is not affected by paraquat at the concentration used here but is affected by H2O2, although to a lesser degree than an nsrR mutation alone. This suggests that overexpression of a member(s) of the NsrR regulon other than hmp also enhances sensitivity to H2O2.
production in the nsrR mutant when Hmp is overexpressed. In vitro, the nsrR hmp double mutant is not affected by paraquat at the concentration used here but is affected by H2O2, although to a lesser degree than an nsrR mutation alone. This suggests that overexpression of a member(s) of the NsrR regulon other than hmp also enhances sensitivity to H2O2.
Acknowledgments
This work was supported by a Research Grant and a postgraduate studentship (N. J. G.) from the Biotechnology and Biological Sciences Research Council (BBSRC, UK). R. C. R. and R. K. P. are recipients of a Wellcome Trust Project Grant (069791/Z/02/Z) which supported important elements of this work. We thank Drs D. Goldberg and D. Monack for strains and J. Green for donation of pTP223. We thank Mr Mark D. Johnson for assistance with the figure preparation.
Abbreviations
Cm, chloramphenicol
FCS, fetal calf serum
GSNO, S-nitrosoglutathione
IFN-γ, interferon-γ
iNOS, inducible nitric oxide synthase
NOS, nitric oxide synthase
Phox, NAD(P)H oxidase
qRT-PCR, quantitative real-time PCR
RNS, reactive nitrogen species
ROS, reactive oxygen species
RT-PCR, reverse transcriptase PCR
SOD, superoxide dismutase
References
- Almeida, C. C., Romao, C. V., Lindley, P. F., Teixeira, M. & Saraiva, L. M. (2006). The role of the hybrid cluster protein in oxidative stress defence. J Biol Chem 281, 32445–32450. [DOI] [PubMed] [Google Scholar]
- Bang, I. S., Liu, L., Vazquez-Torres, A., Crouch, M. L., Stamler, J. S. & Fang, F. C. (2006). Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohaemoglobin Hmp. J Biol Chem 281, 28039–28047. [DOI] [PubMed] [Google Scholar]
- Bodenmiller, D. M. & Spiro, S. (2006). The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol 188, 874–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairrao, F., Cruz, A., Mori, H. & Arraiano, C. M. (2003). Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol Microbiol 50, 1349–1360. [DOI] [PubMed] [Google Scholar]
- Chakravortty, D., Hansen-Wester, I. & Hensel, M. (2002). Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med 195, 1155–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, M. J. & Goldberg, D. E. (1998a). Regulation of the Salmonella typhimurium flavohaemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J Biol Chem 273, 34028–34032. [DOI] [PubMed] [Google Scholar]
- Crawford, M. J. & Goldberg, D. E. (1998b). Role for the Salmonella flavohaemoglobin in protection from nitric oxide. J Biol Chem 273, 12543–12547. [DOI] [PubMed] [Google Scholar]
- Crawford, M. J. & Goldberg, D. E. (2006). Regulation of the Salmonella typhimurium flavohaemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J Biol Chem 281, 3752. [DOI] [PubMed] [Google Scholar]
- Cruz-Ramos, H., Crack, J., Wu, G., Hughes, M. N., Scott, C., Thomson, A. J., Green, J. & Poole, R. K. (2002). NO sensing by FNR: regulation of the Escherichia coli NO-detoxifying flavohaemoglobin, Hmp. EMBO J 21, 3235–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Autreaux, B., Touati, D., Bersch, B., Latour, J.-M. & Michaud-Soret, I. (2002). Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc Natl Acad Sci U S A 99, 16619–16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson, S., Lucchini, S., Thompson, A., Rhen, M. & Hinton, J. C. (2003). Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol 47, 103–118. [DOI] [PubMed] [Google Scholar]
- Fang, F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol 2, 820. [DOI] [PubMed] [Google Scholar]
- Fang, F. & Vazquez-Torres, A. (2002). Salmonella selectively stops traffic. Trends Microbiol 10, 391–392. [DOI] [PubMed] [Google Scholar]
- Fang, F. C., DeGroote, M. A., Foster, J. W., Baumler, A. J., Ochsner, U., Testerman, T., Bearson, S., Giard, J. C., Xu, Y. & other authors (1999). Virulent Salmonella typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc Natl Acad Sci U S A 96, 7502–7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, S. B. & Kogoma, T. (1991). Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev 55, 561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatley, J., Barrett, J., Pullan, S. T., Hughes, M. N., Green, J. & Poole, R. K. (2005). Transcriptional responses of Escherichia coli to S-nitrosoglutathione under defined chemostat conditions reveal major changes in methionine biosynthesis. J Biol Chem 280, 10065–10072. [DOI] [PubMed] [Google Scholar]
- Forman, H. J. & Torres, M. (2002). Reactive oxygen species and cell signalling: respiratory burst in macrophage signalling. Am J Respir Crit Care Med 166, S4–S8. [DOI] [PubMed] [Google Scholar]
- Garcia-del Portillo, F., Foster, J. W. & Finlay, B. B. (1993). Role of acid tolerance response genes in Salmonella typhimurium virulence. Infect Immun 61, 4489–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, P. R., Gardner, A. M., Martin, L. A. & Salzman, A. L. (1998). Nitric oxide dioxygenase: an enzymic function for flavohaemoglobin. Proc Natl Acad Sci U S A 95, 10378–10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell, B. & Gutteridge, J. M. C. (1999). Free Radicals in Biology and Medicine, 3rd edn. Oxford University Press.
- Hart, T. W. (1985). Some observations concerning the S-nitroso and S-phenylsulphonyl derivatives of l-cysteine and glutathione. Tetrahedron Lett 26, 2013–2016. [Google Scholar]
- Hausladen, A., Gow, A. J. & Stamler, J. S. (1998). Nitrosative stress: metabolic pathway involving the flavohaemoglobin. Proc Natl Acad Sci U S A 95, 14100–14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Urzua, E., Zamorano-Sanchez, D. S., Ponce-Coria, J., Morett, E., Grogan, S., Poole, R. K. & Membrillo-Hernandez, J. (2007). Multiple regulators of the flavohaemoglobin (hmp) gene of Salmonella enterica serovar Typhimurium include RamA, a transcriptional regulator conferring the multidrug resistance phenotype. Arch Microbiol 187, 67–77. [DOI] [PubMed] [Google Scholar]
- Ignarro, L. J., Fukuto, J. M., Griscavage, J. M., Rogers, N. E. & Byrns, R. E. (1993). Oxidation of nitric oxide in aqueous solution to nitrite but not nitrate: comparison with enzymatically formed nitric oxide from l-arginine. Proc Natl Acad Sci U S A 90, 8103–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay, J. A. (2002). How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv Microb Physiol 46, 111–153. [DOI] [PubMed] [Google Scholar]
- Justino, M. C., Vicente, J. B., Teixeira, M. & Saraiva, L. M. (2005). New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J Biol Chem 280, 2636–2643. [DOI] [PubMed] [Google Scholar]
- Justino, M. C., Almeida, C. C., Goncalves, V. L., Teixeira, M. & Saraiva, L. M. (2006). Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol Lett 257, 278–284. [DOI] [PubMed] [Google Scholar]
- Kalupahana, R. S., Mastroeni, P., Maskell, D. & Blacklaws, B. A. (2005). Activation of murine dendritic cells and macrophages induced by Salmonella enterica serovar Typhimurium. Immunology 115, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres, D. G., Janakiraman, A., Slauch, J. M. & Maguire, M. E. (2002). Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J Bacteriol 184, 3151–3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonov, V. G., Sundquist, A. R. & Sharma, V. S. (1994). Kinetics of nitric oxide autoxidation in aqueous solution. J Biol Chem 269, 5881–5883. [PubMed] [Google Scholar]
- Kim, C. C., Monack, D. & Falkow, S. (2003). Modulation of virulence by two acidified nitrite-responsive loci of Salmonella enterica serovar Typhimurium. Infect Immun 71, 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liochev, S. I. & Fridovich, I. (1993). Effects of paraquat on Escherichia coli: sensitivity to small changes in pH of the medium – a cautionary note. Arch Biochem Biophys 306, 518–520. [DOI] [PubMed] [Google Scholar]
- Liu, Q. P., Fruit, K., Ward, J. & Correll, P. H. (1999). Negative regulation of macrophage activation in response to IFN-gamma and lipopolysaccharide by the STK/RON receptor tyrosine kinase. J Immunol 163, 6606–6613. [PubMed] [Google Scholar]
- Maloy, S. R., Stewart, V. J. & Taylor, R. K. (1996). Genetic Analysis of Pathogenic Bacteria. A Laboratory Manual. Cold Spring Harbor, USA: Cold Spring Harbor Laboratory Press.
- McCollister, B. D., Bourret, T. J., Gill, R., Jones-Carson, J. & Vazquez-Torres, A. (2005). Repression of SPI2 transcription by nitric oxide-producing, IFNgamma-activated macrophages promotes maturation of Salmonella phagosomes. J Exp Med 202, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Membrillo-Hernandez, J., Ioannidis, N. & Poole, R. K. (1996). The flavohaemoglobin (HMP) of Escherichia coli generates superoxide in vitro and causes oxidative stress in vivo. FEBS Lett 382, 141–144. [DOI] [PubMed] [Google Scholar]
- Membrillo-Hernandez, J., Coopamah, M. D., Channa, A., Hughes, M. N. & Poole, R. K. (1998). A novel mechanism for upregulation of the Escherichia coli K-12 hmp (flavohaemoglobin) gene by the ‘NO releaser’, S-nitrosoglutathione: nitrosation of homocysteine and modulation of MetR binding to the glyA-hmp intergenic region. Mol Microbiol 29, 1101–1112. [DOI] [PubMed] [Google Scholar]
- Membrillo-Hernandez, J., Coopamah, M. D., Anjum, M. F., Stevanin, T. M., Kelly, A., Hughes, M. N. & Poole, R. K. (1999). The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a ‘nitric oxide releaser’, and paraquat and is essential for transcriptional responses to oxidative stress. J Biol Chem 274, 748–754. [DOI] [PubMed] [Google Scholar]
- Mills, C. E., Sedelnikova, S., Soballe, B., Hughes, M. N. & Poole, R. K. (2001). Escherichia coli flavohaemoglobin (Hmp) with equistoichiometric FAD and haem contents has a low affinity for dioxygen in the absence or presence of nitric oxide. Biochem J 353, 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, P., Zheng, M., Bedzyk, L. A., LaRossa, R. A. & Storz, G. (2004). Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci U S A 101, 745–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K. C. (1998). Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol 180, 2063–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, M. M., Geng, H., Nakano, S. & Kobayashi, K. (2006). The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J Bacteriol 188, 5878–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orii, Y., Ioannidis, N. & Poole, R. K. (1992). The oxygenated flavohaemoglobin from Escherichia coli: evidence from photodissociation and rapid-scan studies for two kinetic and spectral forms. Biochem Biophys Res Commun 187, 94–100. [DOI] [PubMed] [Google Scholar]
- Poole, R. K., Anjum, M. F., Membrillo-Hernandez, J., Kim, S. O., Hughes, M. N. & Stewart, V. (1996). Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J Bacteriol 178, 5487–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, R. K., Rogers, N. J., D'mello, R. A., Hughes, M. N. & Orii, Y. (1997). Escherichia coli flavohaemoglobin (Hmp) reduces cytochrome c and Fe(III)-hydroxamate K by electron transfer from NADH via FAD: sensitivity of oxidoreductase activity to haem-bound dioxygen. Microbiology 143, 1557–1565. [DOI] [PubMed] [Google Scholar]
- Poteete, A. R. & Fenton, A. C. (1984). Lambda red-dependent growth and recombination of phage P22. Virology 134, 161–167. [DOI] [PubMed] [Google Scholar]
- Pullan, S. T., Gidley, M. A., Jones, R. A., Barrett, J., Stevanin, T. M., Read, R. C., Green, J. & Poole, R. K. (2007). Nitric oxide in chemostat-cultured Escherichia coli is sensed by fnr and other global regulators; unaltered methionine biosynthesis indicates lack of S-nitrosation. J Bacteriol 189, 1845–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratledge, C. & Dover, L. G. (2000). Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54, 881–941. [DOI] [PubMed] [Google Scholar]
- Read, R. C., Zimmerli, S., Broaddus, C., Sanan, D. A., Stephens, D. S. & Ernst, J. D. (1996). The (α2→8)-linked polysialic acid capsule of group B Neisseria meningitidis modifies multiple steps during interaction with human macrophages. Infect Immun 64, 3210–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renart, J. & Sandoval, I. V. (1984). Western blots. Methods Enzymol 104, 455–460. [DOI] [PubMed] [Google Scholar]
- Rodionov, D. A., Dubchak, I. L., Arkin, A. P., Alm, E. J. & Gelfand, M. S. (2005). Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput Biol 1, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S. & Skaletsky, H. J. (2000). Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press.
- Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Schwartz, C. J., Giel, J. L., Patschkowski, T., Luther, C., Ruzicka, F. J., Beinert, H. & Kiley, P. J. (2001). IscR, an Fe–S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe–S cluster assembly proteins. Proc Natl Acad Sci U S A 98, 14895–14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin, T. M., Ioannidis, N., Mills, C. E., Kim, S. O., Hughes, M. N. & Poole, R. K. (2000). Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo′ or bd, from nitric oxide. J Biol Chem 275, 35868–35875. [DOI] [PubMed] [Google Scholar]
- Stevanin, T. M., Poole, R. K., Demoncheaux, E. A. & Read, R. C. (2002). Flavohemoglobin Hmp protects Salmonella enterica serovar typhimurium from nitric oxide-related killing by human macrophages. Infect Immun 70, 4399–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin, T. M., Moir, J. W. & Read, R. C. (2005). Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infect Immun 73, 3322–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, G. & Imlay, J. A. (1999). Oxidative stress. Curr Opin Microbiol 2, 188–194. [DOI] [PubMed] [Google Scholar]
- Szabo, C., Zingarelli, B., O'Connor, M. & Salzman, A. L. (1996). DNA strand breakage, activation of poly(ADP-ribose) synthetase, and cellular energy depletion are involved in the cytotoxicity in macrophages and smooth muscle cells exposed to peroxynitrite. Proc Natl Acad Sci U S A 93, 1753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe, T., Sasakawa, C., Okada, N., Honma, Y. & Yoshikawa, M. (1992). vacB, a novel chromosomal gene required for expression of virulence genes on the large plasmid of Shigella flexneri. J Bacteriol 174, 6359–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis, R. M., Baumler, A. J. & Heffron, F. (1995). Role of Salmonella typhimurium Mn-superoxide dismutase (SodA) in protection against early killing by J774 macrophages. Infect Immun 63, 1739–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, R. J., Taylor, D. E. & Weiner, J. H. (1997). Expression of Escherichia coli TehA gives resistance to antiseptics and disinfectants similar to that conferred by multidrug resistance efflux pumps. Antimicrob Agents Chemother 41, 440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unanue, E. R. (1993). Macrophages, Antigen-Presenting Cells, and the Phenomena of Antigen Handling and Presentation. New York: Raven Press.
- van den Berg, W. A., Hagen, W. R. & van Dongen, W. M. (2000). The hybrid-cluster protein (‘prismane protein’) from Escherichia coli. Characterization of the hybrid-cluster protein, redox properties of the [2Fe–2S] and [4Fe–2S–2O] clusters and identification of an associated NADH oxidoreductase containing FAD and [2Fe–2S]. Eur J Biochem 267, 666–676. [DOI] [PubMed] [Google Scholar]
- Vasudevan, S. G., Armarego, W. L., Shaw, D. C., Lilley, P. E., Dixon, N. E. & Poole, R. K. (1991). Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol Gen Genet 226, 49–58. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres, A. & Fang, F. C. (2001a). Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol 9, 29–33. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres, A. & Fang, F. C. (2001b). Salmonella evasion of the NADPH phagocyte oxidase. Microbes Infect 3, 1313–1320. [DOI] [PubMed] [Google Scholar]
- Vazquez-Torres, A., Jones-Carson, J., Mastroeni, P., Ischiropoulos, H. & Fang, F. C. (2000). Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med 192, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes-Riesenberg, M. R., Bearson, B., Foster, J. W. & Curtiss, R., III (1996). Role of acid tolerance response in virulence of Salmonella typhimurium. Infect Immun 64, 1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink, D. A., Darbyshire, J. F., Nims, R. W., Saavedra, J. E. & Ford, P. C. (1993). Reactions of the bioregulatory agent nitric oxide in oxygenated aqueous media: determination of the kinetics for oxidation and nitrosation by intermediates generated in the NO/O2 reaction. Chem Res Toxicol 6, 23–27. [DOI] [PubMed] [Google Scholar]
- Witthoft, T., Eckmann, L., Kim, J. M. & Kagnoff, M. F. (1998). Enteroinvasive bacteria directly activate expression of iNOS and NO production in human colon epithelial cells. Am J Physiol 275, G564–G571. [DOI] [PubMed] [Google Scholar]
- Woodmansee, A. N. & Imlay, J. A. (2003). A mechanism by which nitric oxide accelerates the rate of oxidative DNA damage in Escherichia coli. Mol Microbiol 49, 11–22. [DOI] [PubMed] [Google Scholar]
- Wu, G., Wainwright, L. M. & Poole, R. K. (2003). Microbial globins. Adv Microb Physiol 47, 255–310. [DOI] [PubMed] [Google Scholar]
- Wu, G., Corker, H., Orii, Y. & Poole, R. K. (2004). Escherichia coli Hmp, an ‘oxygen-binding flavohaemoprotein’, produces superoxide anion and self-destructs. Arch Microbiol 182, 193–203. [DOI] [PubMed] [Google Scholar]