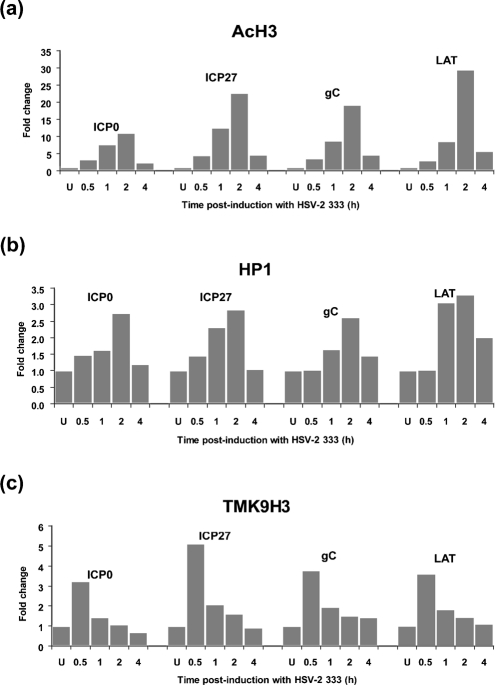
Fig. 4.
ChIP analysis using quantitative real-time PCR. MRC-5 cells were latently infected with in1382 and, at 6 days p.i., either superinfected or not with wt HSV-2 strain 333 for 0.5, 1, 2 or 4 h. Antibodies used were (a) anti-acetyl histone H3 (acetyl K9 and K14) (AcH3), (b) anti-HP1-α (HP1) or (c) anti-trimethyl K9 histone H3 (TMK9H3). PCRs were performed using primer sets to the ICP0, ICP27, gC or LAT promoter. Promoter copy number was quantified by real-time PCR in triplicate and the mean value used to calculate the IP-DNA/input ratio. This value was then expressed as a fold change relative to uninduced material. Fold changes were normalized to GAPDH as described in Methods. Histograms show fold changes in the level of AcH3, HP1 and TMK9H3 at promoters against time post-superinfection with HSV-2. The data shown are representative of three independent experiments.