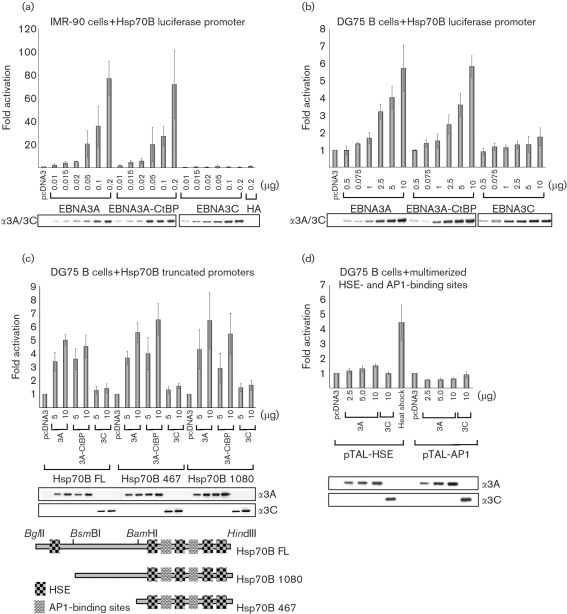
Fig. 7.
EBNA3A and the EBNA3A–CtBP binding mutant activate transcription from an Hsp70B promoter in IMR-90 fibroblasts and DG75 B cells. Truncations of the promoter highlight a 467 bp fragment through which EBNA3A can activate transcription. (a) IMR-90 cells were transfected with 40 ng pGL3-Hsp70B promoter luciferase reporter plasmid and pcDNA3-HA-EBNA3A, pcDNA3-HA-EBNA3A-CtBP binding mutant or pcDNA3-EBNA3C effector plasmid DNA as indicated. Cell extracts were prepared 48 h after transfection and luciferase activity was determined. Data were normalized to β-galactosidase activity (40 ng pSV-βgal per transfection was used) and are expressed as fold activation over pcDNA3 alone (which was given an arbitrary value of 1). Data from three independent experiments are shown, with sd indicated by error bars. (b) DG75 cells were transfected with 2 μg pGL3-Hsp70B luciferase reporter plasmid and effector plasmids as indicated. Means and sd from four independent experiments are shown. Data were normalized to β-galactosidase activity (2 μg pSV-βgal DNA per transfection was used). (c) DG75 cells were transfected as in (b), with two truncated Hsp70B promoter constructs. The truncated promoters and their respective regulatory elements are shown schematically. Means and sd from three independent experiments are shown. (d) EBNA3A has little or no effect on synthetic promoters containing either multimerized HSEs (pTAL-HSE, which was activated by heat shock for 1 h at 43 °C 24 h post-transfection) or AP1-binding sites (pTAL-AP1). Transfections were performed in DG75 B cells and the data from three independent experiments, each including duplicate transfections, are shown. Western blot analysis of one representative experiment is shown for each transfection experiment, indicating that the level of protein expression was comparable for each expression vector.