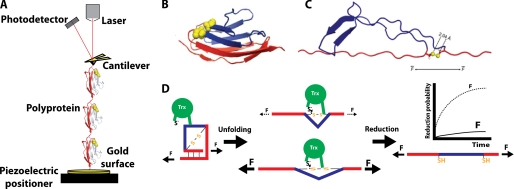
FIGURE 1.
Single-molecule force spectroscopy assay for the detection of single reduction events by the enzyme Trx. A, in the standard assay, (I27G32C-A75C)8 is stretched between the tip of an AFM cantilever and a gold surface. In the force-clamp mode, the signal received by the photodetector is kept constant by an electronic feedback system that controls the extension of the polyprotein via the piezoelectric positioner. Note that, for the sake of simplicity, only three monomers of the polyprotein are depicted in the diagram. B, shown is the I27 monomer, with the residues forming the disulfide bond indicated in yellow (Protein Data Bank code 1TIT). C, shown is the structure of one I27G32C-A75C module after unfolding according to molecular dynamics simulations (27). Because of the presence of the disulfide bond, only part of the protein unfolds (shown in red). D, as long as the modules remain folded, the disulfide bond in each I27G32C-A75C domain cannot be reduced by Trx. The unfolding event acts as a steric switch that allows the disulfide bond to be accommodated into the active site of Trx and be reduced. Concomitant to the reduction, a second length increase is detected, as the trapped residues extend (shown in blue). The probability of reduction drastically depends on the applied force as a consequence of the dynamics of enzyme and substrate during catalysis.